A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study
- PMID: 19138562
- PMCID: PMC2707020
- DOI: 10.1016/S1470-2045(08)70339-5
A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study
Abstract
Background: Genetic subtypes of acute lymphoblastic leukaemia (ALL) are used to determine risk and treatment in children. 25% of precursor B-ALL cases are genetically unclassified and have intermediate prognosis. We aimed to use a genome-wide study to improve prognostic classification of ALL in children.
Methods: We constructed a classifier based on gene expression in 190 children with newly diagnosed ALL (German Cooperative ALL [COALL] discovery cohort) by use of double-loop cross-validation and validated this in an independent cohort of 107 newly diagnosed patients (Dutch Childhood Oncology Group [DCOG] independent validation cohort). Hierarchical cluster analysis with classifying gene-probe sets revealed a new ALL subtype, the underlying genetic abnormalities of which were characterised by comparative genomic hybridisation-arrays and molecular cytogenetics.
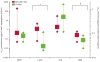
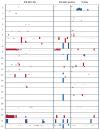
Findings: Our classifier predicted ALL subtype with a median accuracy of 90.0% (IQR 88.3-91.7) in the discovery cohort and correctly identified 94 of 107 patients (accuracy 87.9%) in the independent validation cohort. Without our classifier, 44 children in the COALL cohort and 33 children in the DCOG cohort would have been classified as B-other. However, hierarchical clustering showed that many of these genetically unclassified cases clustered with BCR-ABL1-positive cases: 30 (19%) of 154 children with precursor B-ALL in the COALL cohort and 14 (15%) of 92 children with precursor B-ALL in the DCOG cohort had this BCR-ABL1-like disease. In the COALL cohort, these patients had unfavourable outcome (5-year disease-free survival 59.5%, 95% CI 37.1-81.9) compared with patients with other precursor B-ALL (84.4%, 76.8-92.1%; p=0.012), a prognosis similar to that of patients with BCR-ABL1-positive ALL (51.9%, 23.1-80.6%). In the DCOG cohort, the prognosis of BCR-ABL1-like disease (57.1%, 31.2-83.1%) was worse than that of other precursor B-ALL (79.2%, 70.2-88.3%; p=0.026), and similar to that of BCR-ABL1-positive ALL (32.5%, 2.3-62.7%). 36 (82%) of the patients with BCR-ABL1-like disease had deletions in genes involved in B-cell development, including IKZF1, TCF3, EBF1, PAX5, and VPREB1; only nine (36%) of 25 patients with B-other ALL had deletions in these genes (p=0.0002). Compared with other precursor B-ALL cells, BCR-ABL1-like cells were 73 times more resistant to L-asparaginase (p=0.001) and 1.6 times more resistant to daunorubicin (p=0.017), but toxicity of prednisolone and vincristine did not differ.
Interpretation: New treatment strategies are needed to improve outcome for this newly identified high-risk subtype of ALL.
Funding: Dutch Cancer Society, Sophia Foundation for Medical Research, Paediatric Oncology Foundation Rotterdam, Centre of Medical Systems Biology of the Netherlands Genomics Initiative/Netherlands Organisation for Scientific Research, American National Institute of Health, American National Cancer Institute, and American Lebanese Syrian Associated Charities.
Conflict of interest statement
The authors do not report conflict of interests related to this paper except the submission of a patent application (PCT/NL08/050373) for classification of leukemia by gene expression signatures by MLDB and RP.
Figures


Comment in
-
A new subgroup of high-risk acute lymphoblastic leukaemia.Lancet Oncol. 2009 Feb;10(2):101-3. doi: 10.1016/S1470-2045(09)70008-7. Lancet Oncol. 2009. PMID: 19185828 No abstract available.
References
-
- Pieters R, Schrappe M, De Lorenzo P, et al. A treatment protocol for infants younger than one year of age with acute lymphoblastic leukemia (Interfant-99): an observational study and multicentre randomised trial. Lancet. 2007;370:240–50. - PubMed
-
- Pui CH, Relling MV, Downing JR. Acute lymphoblastic leukemia. N Engl J Med. 2004;350:1535–48. - PubMed
-
- Pieters R, Carroll WL. Biology and treatment of acute lymphoblastic leukemia. Pediatr Clin North Am. 2008;55:1–20. - PubMed
-
- Pui CH, Raimondi SC, Hancock ML, et al. Immunologic, cytogenetic and clinical characterization of childhood acute lymphoblastic leukemia with the t(1;19)(q23;p13) or its derivative. J Clin Oncol. 1994;12:2601–6. - PubMed
-
- Möricke A, Reiter A, Zimmermann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111:4477–89. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

