Duplication within the SEPT9 gene associated with a founder effect in North American families with hereditary neuralgic amyotrophy
- PMID: 19139049
- PMCID: PMC2722193
- DOI: 10.1093/hmg/ddp014
Duplication within the SEPT9 gene associated with a founder effect in North American families with hereditary neuralgic amyotrophy
Abstract
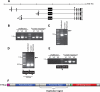
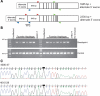
Hereditary neuralgic amyotrophy (HNA) is an autosomal dominant disorder associated with recurrent episodes of focal neuropathy primarily affecting the brachial plexus. Point mutations in the SEPT9 gene have been previously identified as the molecular basis of HNA in some pedigrees. However in many families, including those from North America demonstrating a genetic founder haplotype, no sequence mutations have been detected. We report an intragenic 38 Kb SEPT9 duplication that is linked to HNA in 12 North American families that share the common founder haplotype. Analysis of the breakpoints showed that the duplication is identical in all pedigrees, and molecular analysis revealed that the duplication includes the 645 bp exon in which previous HNA mutations were found. The SEPT9 transcript variants that span this duplication contain two in-frame repeats of this exon, and immunoblotting demonstrates larger molecular weight SEPT9 protein isoforms. This exon also encodes for a majority of the SEPT9 N-terminal proline rich region suggesting that this region plays a role in the pathogenesis of HNA.
Figures

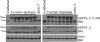



References
-
- Kuhlenbäumer G., Stögbauer F., Timmerman V., De Jonghe P. Diagnostic guidelines for hereditary neuralgic amyotrophy or heredofamilial neuritis with brachial plexus predilection. On behalf of the European CMT Consortium. Neuromuscul. Disord. 2000;10:515–517. - PubMed
-
- Hannibal M.C., van Alfen N., Chance P.F., van Engelen B.G. GeneReviews at GeneTests: Medical Genetics Information Resource. Seattle: Copyright, University of Washington; 2008. pp. 1997–2008. Available at http://www.genetests.org .
-
- van Alfen N., van Engelen B.G. The clinical spectrum of neuralgic amyotrophy in 246 cases. Brain. 2006;129:438–450. - PubMed
-
- Jeannet P.Y., Watts G.D., Bird T.D., Chance P.F. Craniofacial and cutaneous findings expand the phenotype of hereditary neuralgic amyotrophy. Neurology. 2001;57:1963–1968. - PubMed
-
- Gardner J.H., Maloney W. Hereditary brachial and cranial neuritis genetically linked with ocular hypotelorism and syndactyly. Neurology. 1968;18:278. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

