Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay
- PMID: 19139134
- PMCID: PMC2676116
- DOI: 10.1158/1535-7163.MCT-08-0878
Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay
Abstract
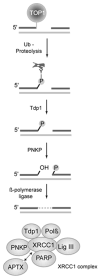
Tyrosyl-DNA phosphodiesterase I (Tdp1) resolves topoisomerase I (Top1)-DNA adducts accumulated from natural DNA damage as well as from the action of certain anticancer drugs. Tdp1 catalyzes the hydrolysis of the phosphodiester bond between the catalytic tyrosine residue of topoisomerase I and the DNA 3'-phosphate. Only a limited number of weak inhibitors have been reported for Tdp1, and there is an unmet need to identify novel chemotypes through screening of chemical libraries. Herein, we present an easily configured, highly miniaturized, and robust Tdp1 assay using the AlphaScreen technology. Uninhibited enzyme reaction is associated with low signal, whereas inhibition leads to a gain of signal, making the present assay format especially attractive for automated large-collection high-throughput screening. We report the identification and initial characterization of four previously unreported inhibitors of Tdp1. Among them, suramin, NF449, and methyl-3,4-dephostatin are phosphotyrosine mimetics that may act as Tdp1 substrate decoys. We also report a novel biochemical assay using the SCAN1 Tdp1 mutant to study the mechanism of action of methyl-3,4-dephostatin.
Figures





References
-
- Pommier Y, Redon C, Rao A, Seiler JA, Sordet O, Takemura H, Antony S, Meng LH, Liao ZY, Kohlhagen G, et al. Repair of and Checkpoint Response to Topoisomerase I-Mediated DNA Damage. Mutat Res. 2003;532:173–203. - PubMed
-
- Pouliot JJ, Yao KC, Robertson CA, Nash HA. Yeast gene for a Tyr-DNA phosphodiesterase that repairs topo I covalent complexes. Science. 1999;286:552–555. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

