A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground
- PMID: 19139297
- PMCID: PMC4111150
- DOI: 10.1001/archneurol.2008.540
A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground
Abstract
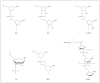
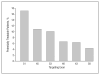
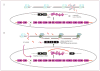
Antisense oligonucleotides are short nucleic acid sequences designed for use as small-molecule drugs. They recognize and bind to specific messenger RNA (mRNA) or pre-mRNA sequences to create small double-stranded regions of the target mRNA that alter mRNA splicing patterns or inhibit protein translation. Antisense approaches have been actively pursued as a form of molecular medicine for more than 20 years, but only one has been translated to a marketed drug (intraocular human immunodeficiency virus treatment). Two recent advances foreshadow a change in clinical applications of antisense strategies. First is the development of synthetic DNA analogues that show outstanding stability and sequence specificity yet little or no binding to modulator proteins. Second is the publication of impressive preclinical and clinical data using antisense in an exon-skipping strategy to increase dystrophin production in Duchenne muscular dystrophy. As long-standing barriers are successfully circumvented, attention turns toward scale-up of production, long-term toxicity studies, and the challenges to traditional drug regulatory attitudes presented by tightly targeted sequence-specific drugs.
Figures

References
-
- Gitlin L, Karelsky S, Andino R. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature. 2002;418(6896):430–434. - PubMed
-
- Kim SK, Wold BJ. Stable reduction of thymidine kinase activity in cells expressing high levels of anti-sense RNA. Cell. 1985;42(1):129–138. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

