Optimization of gene expression by natural selection
- PMID: 19139403
- PMCID: PMC2633540
- DOI: 10.1073/pnas.0812009106
Optimization of gene expression by natural selection
Abstract
It is generally assumed that stabilizing selection promoting a phenotypic optimum acts to shape variation in quantitative traits across individuals and species. Although gene expression represents an intensively studied molecular phenotype, the extent to which stabilizing selection limits divergence in gene expression remains contentious. In this study, we present a theoretical framework for the study of stabilizing and directional selection using data from between-species divergence of continuous traits. This framework, based upon Brownian motion, is analytically tractable and can be used in maximum-likelihood or Bayesian parameter estimation. We apply this model to gene-expression levels in 7 species of Drosophila, and find that gene-expression divergence is substantially curtailed by stabilizing selection. However, we estimate the selective effect, s, of gene-expression change to be very small, approximately equal to Ns for a change of one standard deviation, where N is the effective population size. These findings highlight the power of natural selection to shape phenotype, even when the fitness effects of mutations are in the nearly neutral range.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
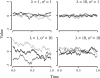
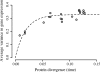
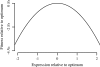
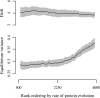
References
-
- Smith NG, Eyre-Walker A. Adaptive protein evolution in Drosophila. Nature. 2002;415:1022–1024. - PubMed
-
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. - PubMed
-
- Carroll SB, Grenier JK, Weatherbee SD. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. New York: Blackwell; 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

