Structure and membrane interaction of myristoylated ARF1
- PMID: 19141284
- PMCID: PMC2659477
- DOI: 10.1016/j.str.2008.10.020
Structure and membrane interaction of myristoylated ARF1
Abstract
ADP-ribosylation factors (ARFs) are small (21 kDa), monomeric GTPases that are important regulators of membrane traffic. When membrane bound, they recruit soluble adaptors to membranes and trigger the assembly of coating complexes involved in cargo selection and vesicular budding. N-myristoylation is a conserved feature of all ARF proteins that is required for its biological functions, although the mechanism(s) by which the myristate acts in ARF functions is not fully understood. Here we present the structure of a myristoylated ARF1 protein, determined by solution NMR methods, and an assessment of the influence of myristoylation on association of ARF1.GDP and ARF1.GTP with lipid bilayers. A model in which myristoylation contributes to both the regulation of guanine nucleotide exchange and stable membrane association is supported.
Figures
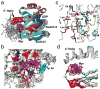

Comment in
-
Journey to the ends of the Arf.Structure. 2009 Jan 14;17(1):2-4. doi: 10.1016/j.str.2008.12.002. Structure. 2009. PMID: 19141275 No abstract available.
References
-
- Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature. 1994;372:704–708. - PubMed
-
- Amor JC, Horton JR, Zhu X, Wang Y, Sullards C, Ringe D, Cheng X, Kahn RA. Structures of yeast ARF2 and ARL1: distinct roles for the N terminus in the structure and function of ARF family GTPases. J Biol Chem. 2001;276:42477–42484. - PubMed
-
- Bigay J, Gounon P, Robineau S, Antonny B. Lipid packing sensed by ArfGAP1 couples COPI coat disassembly to membrane bilayer curvature. Nature. 2003;426:563–566. - PubMed
-
- Bonifacino JS. The GGA proteins: Adaptors on the move. Nature Reviews Molecular Cell Biology. 2004;5:23–32. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

