Riboswitch RNAs: using RNA to sense cellular metabolism
- PMID: 19141470
- PMCID: PMC3959987
- DOI: 10.1101/gad.1747308
Riboswitch RNAs: using RNA to sense cellular metabolism
Abstract
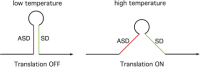
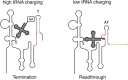
Riboswitches are RNA elements that undergo a shift in structure in response to binding of a regulatory molecule. These elements are encoded within the transcript they regulate, and act in cis to control expression of the coding sequence(s) within that transcript; their function is therefore distinct from that of small regulatory RNAs (sRNAs) that act in trans to regulate the activity of other RNA transcripts. Riboswitch RNAs control a broad range of genes in bacterial species, including those involved in metabolism or uptake of amino acids, cofactors, nucleotides, and metal ions. Regulation occurs as a consequence of direct binding of an effector molecule, or through sensing of a physical parameter such as temperature. Here we review the global role of riboswitch RNAs in bacterial cell metabolism.
Figures

References
-
- Babitzke P., Romeo T. CsrB sRNA family: Sequestration of RNA-binding regulatory proteins. Curr. Opin. Microbiol. 2007;10:156–163. - PubMed
-
- Batey R.T., Gilbert S.D., Montange R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
