Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional
- PMID: 19141533
- PMCID: PMC2669427
- DOI: 10.1096/fj.08-122903
Nuclear reprogramming in heterokaryons is rapid, extensive, and bidirectional
Abstract
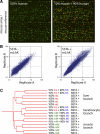
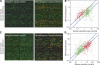
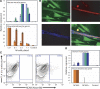
An understanding of nuclear reprogramming is fundamental to the use of cells in regenerative medicine. Due to technological obstacles, the time course and extent of reprogramming of cells following fusion has not been assessed to date. Here, we show that hundreds of genes are activated or repressed within hours of fusion of human keratinocytes and mouse muscle cells in heterokaryons, and extensive changes are observed within 4 days. This study was made possible by the development of a broadly applicable approach, species-specific transcriptome amplification (SSTA), which enables global resolution of transcripts derived from the nuclei of two species, even when the proportions of species-specific transcripts are highly skewed. Remarkably, either phenotype can be dominant; an excess of primary keratinocytes leads to activation of the keratinocyte program in muscle cells and the converse is true when muscle cells are in excess. We conclude that nuclear reprogramming in heterokaryons is rapid, extensive, bidirectional, and dictated by the balance of regulators contributed by the cell types.
Figures



References
-
- Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr Biol. 2001;11:1553–1558. - PubMed
-
- Cowan C A, Atienza J, Melton D A, Eggan K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science. 2005;309:1369–1373. - PubMed
-
- Silva J, Chambers I, Pollard S, Smith A. Nanog promotes transfer of pluripotency after cell fusion. Nature. 2006;441:997–1001. - PubMed
-
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. - PubMed
-
- Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

