Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes
- PMID: 19141596
- PMCID: PMC2652205
- DOI: 10.1101/gr.084038.108
Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes
Abstract
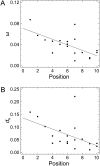
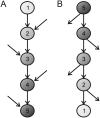
Biological function is based on complex networks consisting of large numbers of interacting molecules. The evolutionary properties of molecular networks and, in particular, the impact of network architecture on the sequence evolution of its individual components are, nonetheless, still poorly understood. Here, we conducted a fine-scale network-level molecular evolutionary analysis of the insulin/TOR pathway across 12 species of Drosophila. We found that the insulin/TOR pathway components are completely conserved across these species and that two genes located at major network branch points show evidence for positive selection. Remarkably, we detected a gradient in the strength of purifying selection along the pathway, increasing from the upstream to the downstream genes. We also found that physically interacting proteins tend to have more similar levels of selective constraint, even though this feature might represent a byproduct of the correlation between selective constraint and the pathway position. Our results clearly indicate that the levels of functional constraint do depend on the position of the proteins in the pathway and, consequently, the architecture of the pathway constrains gene sequence evolution.
Figures



References
-
- Adams M.D., Celniker S.E., Holt R.A., Evans C.A., Gocayne J.D., Amanatides P.G., Scherer S.E., Li P.W., Hoskins R.A., Galle R.F., et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–2195. - PubMed
-
- Avruch J., Belham C., Weng Q., Hara K., Yonezawa K. The p70 S6 kinase integrates nutrient and growth signals to control translational capacity. Prog. Mol. Subcell. Biol. 2001;26:115–154. - PubMed
-
- Begun D.J., Holloway A.K., Stevens K., Hillier L.W., Poh Y.P., Hahn M.W., Nista P.M., Jones C.D., Kern A.D., Dewey C.N., et al. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans . PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. - DOI - PMC - PubMed
-
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300.
-
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000;17:540–552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
