Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy
- PMID: 19141597
- PMCID: PMC2652209
- DOI: 10.1101/gr.075671.107
Genomic copy number determination in cancer cells from single nucleotide polymorphism microarrays based on quantitative genotyping corrected for aneuploidy
Erratum in
- Genome Res. 2009 Mar;19(3):520
Abstract
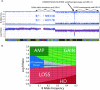
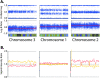
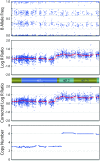
Microarrays are frequently used to profile genome-wide copy number (CN) aberrations. While generally robust for detecting CN variants in germline DNA, the methods used to derive CN from signal intensity values have been suboptimal when applied to cancer genomes. The complexity of genomic aberrations in cancer makes it more difficult to discriminate between signal and noise, and measuring CN as a discrete variable does not account for tumor heterogeneity. Furthermore, standard normalization approaches detect CN changes relative to the overall DNA content, which is often not diploid in cancer. We propose an algorithm that uses the degree of allelic imbalance as well as probe intensity, with a correction for aneuploidy, for a quantitative CN assessment and scoring of allelic ratios. This algorithm results in a more precise definition of CN and allelic aberration in the cancer genome, which is essential for translational efforts focused on using these tools for molecular diagnostics and for the discovery of therapeutic targets.
Figures



References
-
- Albertson D.G., Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12:R145–R152. - PubMed
-
- Attiyeh E.F., London W.B., Mosse Y.P., Wang Q., Winter C., Khazi D., McGrady P.W., Seeger R.C., Look A.T., Shimada H., et al. Chromosome 1p and 11q deletions and outcome in neuroblastoma. N. Engl. J. Med. 2005;353:2243–2253. - PubMed
-
- Fan J.B., Chee M.S., Gunderson K.L. Highly parallel genomic assays. Nat. Rev. Genet. 2006;7:632–644. - PubMed
-
- George R., Attiyeh E., Li S., Neuberg D., Li C., Hii G., Fox E., Meyerson M., Look A., Maris J. Genome-wide analysis of neuroblastomas using high-density single nucleotide polymorphism (SNP) arrays. American Association for Cancer Research Annual Meeting; Anaheim, CA. 2005.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
