Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resouce-limited West Africa
- PMID: 19143530
- PMCID: PMC3671065
- DOI: 10.1086/596504
Emergence of multiclass drug-resistance in HIV-2 in antiretroviral-treated individuals in Senegal: implications for HIV-2 treatment in resouce-limited West Africa
Erratum in
- Clin Infect Dis. 2009 Mar 15;48(6):848
Abstract
Background: The efficacy of various antiretroviral (ARV) therapy regimens for human immunodeficiency virus type 2 (HIV-2) infection remains unclear. HIV-2 is intrinsically resistant to the nonnucleoside reverse-transcriptase inhibitors and to enfuvirtide and may also be less susceptible than HIV-1 to some protease inhibitors (PIs). However, the mutations in HIV-2 that confer ARV resistance are not well characterized.
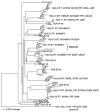
Methods: Twenty-three patients were studied as part of an ongoing prospective longitudinal cohort study of ARV therapy for HIV-2 infection in Senegal. Patients were treated with nucleoside reverse-transcriptase inhibitor (NRTI)- and PI (indinavir)-based regimens. HIV-2 pol genes from these patients were genotyped, and the mutations predictive of resistance in HIV-2 were assessed. Correlates of ARV resistance were analyzed.
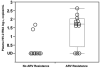
Results: Multiclass drug-resistance mutations (NRTI and PI) were detected in strains in 30% of patients; 52% had evidence of resistance to at least 1 ARV class. The reverse-transcriptase mutations M184V and K65R, which confer high-level resistance to lamivudine and emtricitabine in HIV-2, were found in strains from 43% and 9% of patients, respectively. The Q151M mutation, which confers multinucleoside resistance in HIV-2, emerged in strains from 9% of patients. HIV-1-associated thymidine analogue mutations (M41L, D67N, K70R, L210W, and T215Y/F) were not observed, with the exception of K70R, which was present together with K65R and Q151M in a strain from 1 patient. Eight patients had HIV-2 with PI mutations associated with indinavir resistance, including K7R, I54M, V62A, I82F, L90M, L99F; 4 patients had strains with multiple PI resistance-associated mutations. The duration of ARV therapy was positively associated with the development of drug resistance (P = .02). Nine (82%) of 11 patients with HIV-2 with no [corrected] detectable ARV resistance had undetectable plasma HIV-2 RNA loads (<1.4 log(10) copies/mL), compared with 3 (25%) of 12 patients with HIV-2 with detectable ARV resistance (P = .009). Patients with ARV-resistant virus had higher plasma HIV-2 RNA loads, compared with those with non-ARV-resistant virus (median, 1.7 log(10) copies/mL [range, <1.4 to 2.6 log(10) copies/mL] vs. <1.4 log(10) copies/mL [range, <1.4 to 1.6 log(10) copies/mL]; P = .003).
Conclusions: HIV-2-infected individuals treated with ARV therapy in Senegal commonly have HIV-2 mutations consistent with multiclass drug resistance. Additional clinical studies are required to improve the efficacy of primary and salvage treatment regimens for treating HIV-2 infection.
Conflict of interest statement
Figures
References
-
- De Cock KM, Adjorlolo G, Ekpini E, et al. Epidemiology and transmission of HIV-2: why there is no HIV-2 pandemic. JAMA. 1993;270:2083–6. - PubMed
-
- Simon F, Matheron S, Tamalet C, et al. Cellular and plasma viral load in patients infected with HIV-2. AIDS. 1993;7:1411–7. - PubMed
-
- Marlink R, Kanki P, Thior I, et al. Reduced rate of disease development after HIV-2 infection as compared to HIV-1. Science. 1994;265:1587–90. - PubMed
-
- Kanki PJ, Travers KU, MBoup S, et al. Slower heterosexual spread of HIV-2 than HIV-1. Lancet. 1994;343:943–6. - PubMed
-
- Gottlieb GS, Sow PS, Hawes SE, et al. Equal plasma viral loads predict a similar rate of CD4+ T cell decline in human immunodeficiency virus (HIV) type 1– and HIV-2–infected individuals from Senegal, West Africa. J Infect Dis. 2002;185:905–14. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

