Biofilm formation by Staphylococcus haemolyticus
- PMID: 19144798
- PMCID: PMC2668337
- DOI: 10.1128/JCM.01891-08
Biofilm formation by Staphylococcus haemolyticus
Abstract
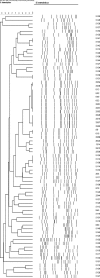
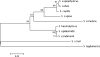
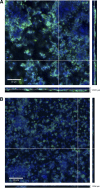
Infections due to coagulase-negative staphylococci (CoNS) most frequently occur after the implantation of medical devices and are attributed to the biofilm-forming potential of CoNS. Staphylococcus haemolyticus is the second most frequently isolated CoNS from patients with hospital-acquired infections. There is only limited knowledge of the nature of S. haemolyticus biofilms. The aim of this study was to characterize S. haemolyticus biofilm formation. We analyzed the biofilm-forming capacities of 72 clinical S. haemolyticus isolates. A detachment assay with NaIO(4), proteinase K, or DNase was used to determine the main biofilm components. Biofilm-associated genes, including the ica operon, were analyzed by PCR, and the gene products were sequenced. Confocal laser scanning microscopy (CLSM) was used to elucidate the biofilm structure. Fifty-three isolates (74%) produced biofilms after growth in Trypticase soy broth (TSB) with glucose, but only 22 (31%) produced biofilms after growth in TSB with NaCl. It was necessary to dissolve the biofilm in ethanol-acetone to measure the optical density of the full biofilm mass. DNase, proteinase K, and NaIO(4) caused biofilm detachment for 100%, 98%, and 38% of the isolates, respectively. icaRADBC and polysaccharide intercellular adhesin (PIA) production were found in only two isolates. CLSM indicated that the biofilm structure of S. haemolyticus clearly differs from that of S. epidermidis. We conclude that biofilm formation is a common phenotype in clinical S. haemolyticus isolates. In contrast to S. epidermidis, proteins and extracellular DNA are of functional relevance for biofilm accumulation, whereas PIA plays only a minor role. The induction of biofilm formation and determination of the biofilm mass also needed to be optimized for S. haemolyticus.
Figures


References
-
- Al Laham, N., H. Rohde, G. Sander, A. Fischer, M. Hussain, C. Heilmann, D. Mack, R. Proctor, G. Peters, K. Becker, and C. von Eiff. 2007. Augmented expression of polysaccharide intercellular adhesin in a defined Staphylococcus epidermidis mutant with the small-colony-variant phenotype. J. Bacteriol. 1894494-4501. - PMC - PubMed
-
- Anonymous. 1999. National Nosocomial Infections Surveillance System report: data summary from January 1990-May 1999. National Nosocomial Infections Surveillance System, Centers for Disease Control and Prevention, Atlanta, GA.
-
- Arbeidsgruppen for Antibiotikaspørsmål. 2008. NWGA breakpoints for susceptibilities to bacteria, version 1.11. Arbeidsgruppen for Antibiotikaspørsmål, Oslo, Norway.
-
- Barton, B. M., G. P. Harding, and A. J. Zuccarelli. 1995. A general method for detecting and sizing large plasmids. Anal. Biochem. 226235-240. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

