The proteasome: overview of structure and functions
- PMID: 19145068
- PMCID: PMC3524306
- DOI: 10.2183/pjab.85.12
The proteasome: overview of structure and functions
Abstract
The proteasome is a highly sophisticated protease complex designed to carry out selective, efficient and processive hydrolysis of client proteins. It is known to collaborate with ubiquitin, which polymerizes to form a marker for regulated proteolysis in eukaryotic cells. The highly organized proteasome plays a prominent role in the control of a diverse array of basic cellular activities by rapidly and unidirectionally catalyzing biological reactions. Studies of the proteasome during the past quarter of a century have provided profound insights into its structure and functions, which has appreciably contributed to our understanding of cellular life. Many questions, however, remain to be elucidated.
Figures
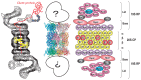


References
-
- Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 - PubMed
-
- Varshavsky A. (2005) Regulated protein degradation. Trends Biochem. Sci. 30, 283–286 - PubMed
-
- Ciechanover A. (2006) The ubiquitin proteolytic system: from a vague idea, through basic mechanisms, and onto human diseases and drug targeting. Neurology 66, S7–19 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

