Establishing new sites of polarization by microtubules
- PMID: 19147354
- PMCID: PMC2820583
- DOI: 10.1016/j.cub.2008.12.008
Establishing new sites of polarization by microtubules
Abstract
Background: Microtubules (MTs) participate in the spatial regulation of actin-based processes such as cytokinesis and cell polarization. The fission yeast Schizosaccharomyces pombe is a rod-shaped cell that exhibits polarized cell growth at cell tips. MT plus ends contact and shrink from the cell tips and contribute to polarity regulation.
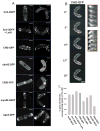
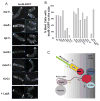
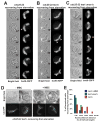
Results: Here, we investigate the effects of changing cell shape on MTs and cell-polarization machinery. We physically bend fission yeast cells by forcing them into microfabricated femtoliter chambers. In these bent cells, MTs maintain a straight axis and contact and shrink from cortical sites at the sides of cells. At these ectopic sites, polarity factors such as bud6p, for3p (formin), and cdc42p are recruited and assemble actin cables in a MT-dependent manner. MT contact at the cortex induces the appearance of a bud6p dot within seconds. The accumulation of polarity factors leads to cell growth at these sites, when the MT-associated polarity factor tea1p is absent. This process is dependent on MTs, mal3p (EB1), moe1p (an EB1-binding protein), and for3p but, surprisingly, is independent of the tea1p-tea4p pathway.
Conclusions: These studies provide a direct demonstration for how MTs induce actin assembly at specific locations on the cell cortex and begin to identify a new pathway involved in this process. MT interactions with the cortex may be regulated by cortical-attachment sites. These findings highlight the crosstalk between cell shape, polarity mechanisms, and MTs responsible for cell morphogenesis.
Figures



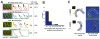
Comment in
-
Cell polarization: it's all about being in shape.Curr Biol. 2009 Mar 10;19(5):R205-6. doi: 10.1016/j.cub.2009.01.024. Curr Biol. 2009. PMID: 19278635
Similar articles
-
Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity.Dev Cell. 2005 Apr;8(4):479-91. doi: 10.1016/j.devcel.2005.02.008. Dev Cell. 2005. PMID: 15809031
-
Regulation of actin assembly by microtubules in fission yeast cell polarity.Novartis Found Symp. 2005;269:59-66; discussion 66-72, 223-30. Novartis Found Symp. 2005. PMID: 16355535
-
Role of bud6p and tea1p in the interaction between actin and microtubules for the establishment of cell polarity in fission yeast.Curr Biol. 2001 Jun 5;11(11):836-45. doi: 10.1016/s0960-9822(01)00235-4. Curr Biol. 2001. PMID: 11516644
-
Shaping fission yeast with microtubules.Cold Spring Harb Perspect Biol. 2009 Jul;1(1):a001347. doi: 10.1101/cshperspect.a001347. Cold Spring Harb Perspect Biol. 2009. PMID: 20066076 Free PMC article. Review.
-
Cell shape and cell division in fission yeast.Curr Biol. 2009 Sep 15;19(17):R823-7. doi: 10.1016/j.cub.2009.08.012. Curr Biol. 2009. PMID: 19906584 Free PMC article. Review.
Cited by
-
End-on microtubule-dynein interactions and pulling-based positioning of microtubule organizing centers.Cell Cycle. 2012 Oct 15;11(20):3750-7. doi: 10.4161/cc.21753. Epub 2012 Aug 16. Cell Cycle. 2012. PMID: 22895049 Free PMC article. Review.
-
Kinesin-4 optimizes microtubule orientations for responsive tip growth guidance in moss.J Cell Biol. 2023 Sep 4;222(9):e202202018. doi: 10.1083/jcb.202202018. Epub 2023 Jun 30. J Cell Biol. 2023. PMID: 37389658 Free PMC article.
-
Reliable and robust control of nucleus centering is contingent on nonequilibrium force patterns.iScience. 2023 Apr 13;26(5):106665. doi: 10.1016/j.isci.2023.106665. eCollection 2023 May 19. iScience. 2023. PMID: 37182105 Free PMC article.
-
Influence of cell geometry on division-plane positioning.Cell. 2011 Feb 4;144(3):414-26. doi: 10.1016/j.cell.2011.01.016. Cell. 2011. PMID: 21295701 Free PMC article.
-
Fabrication of microcompartments with controlled size and shape for encapsulating active matter.Front Cell Dev Biol. 2025 Jun 20;13:1616089. doi: 10.3389/fcell.2025.1616089. eCollection 2025. Front Cell Dev Biol. 2025. PMID: 40620762 Free PMC article.
References
-
- Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. - PubMed
-
- Wen Y, Eng CH, Schmoranzer J, Cabrera-Poch N, Morris EJ, Chen M, Wallar BJ, Alberts AS, Gundersen GG. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat Cell Biol. 2004;6:820–830. - PubMed
-
- Martin SG, Chang F. New end take off: regulating cell polarity during the fission yeast cell cycle. Cell Cycle. 2005;4:1046–1049. - PubMed
-
- Brunner D, Nurse P. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell. 2000;102:695–704. - PubMed
-
- Chang F, Peter M. Yeasts make their mark. Nat Cell Biol. 2003;5:294–299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

