Synaptic Vesicle Protein NTT4/XT1 (SLC6A17) Catalyzes Na+-coupled Neutral Amino Acid Transport
- PMID: 19147495
- PMCID: PMC2659202
- DOI: 10.1074/jbc.M806407200
Synaptic Vesicle Protein NTT4/XT1 (SLC6A17) Catalyzes Na+-coupled Neutral Amino Acid Transport
Abstract
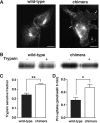
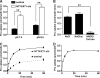
The SLC6 family of structurally related, Na(+)-dependent transporter proteins is responsible for presynaptic reuptake of the majority of neurotransmitters. Within this family are a number of orphan transporters, including NTT4/XT1 (SLC6A17), a protein first identified over 15 years ago. NTT4/XT1 is expressed exclusively in the nervous system and specifically on synaptic vesicles in glutamatergic and some GABAergic neurons. Despite extensive efforts by a number of groups, no substrate has been reported for NTT4/XT1. Here we use a combination of molecular manipulations to increase expression of the NTT4/XT1 protein at the plasma membrane and to directly demonstrate that it catalyzes neutral amino acid transport. The substrate profile of the NTT4/XT1-dependent activity is similar to that of the closely related B(0)AT2/SBAT1 (SLC6A15), including a submillimolar apparent affinity for proline and leucine and a low millimolar apparent affinity for glutamine. The transport activity is Na(+)-dependent and Cl(-)-independent and is inhibited by low pH as is SLC6A15, suggesting redundant roles for these proteins. This characterization of NTT4/XT1 offers important insights into neurotransmitter metabolism as well as the mechanistic differences among the structurally related, but functionally divergent, SLC6 proteins.
Figures




Similar articles
-
The orphan transporter Rxt1/NTT4 (SLC6A17) functions as a synaptic vesicle amino acid transporter selective for proline, glycine, leucine, and alanine.Mol Pharmacol. 2008 Dec;74(6):1521-32. doi: 10.1124/mol.108.050005. Epub 2008 Sep 3. Mol Pharmacol. 2008. PMID: 18768736
-
The orphan transporter v7-3 (slc6a15) is a Na+-dependent neutral amino acid transporter (B0AT2).Biochem J. 2006 Jan 1;393(Pt 1):421-30. doi: 10.1042/BJ20051273. Biochem J. 2006. PMID: 16185194 Free PMC article.
-
Characterization of a branched-chain amino-acid transporter SBAT1 (SLC6A15) that is expressed in human brain.Biochem Biophys Res Commun. 2005 Nov 25;337(3):892-900. doi: 10.1016/j.bbrc.2005.09.128. Epub 2005 Sep 30. Biochem Biophys Res Commun. 2005. PMID: 16226721
-
The SLC6 orphans are forming a family of amino acid transporters.Neurochem Int. 2006 May-Jun;48(6-7):559-67. doi: 10.1016/j.neuint.2005.11.021. Epub 2006 Mar 15. Neurochem Int. 2006. PMID: 16540203 Review.
-
The SLC6A15-SLC6A20 Neutral Amino Acid Transporter Subfamily: Functions, Diseases, and Their Therapeutic Relevance.Pharmacol Rev. 2023 Dec 15;76(1):142-193. doi: 10.1124/pharmrev.123.000886. Pharmacol Rev. 2023. PMID: 37940347 Review.
Cited by
-
Chloride-dependent conformational changes in the GlyT1 glycine transporter.Proc Natl Acad Sci U S A. 2021 Mar 9;118(10):e2017431118. doi: 10.1073/pnas.2017431118. Proc Natl Acad Sci U S A. 2021. PMID: 33658361 Free PMC article.
-
A novel role for PHT1 in the disposition of l-histidine in brain: In vitro slice and in vivo pharmacokinetic studies in wildtype and Pht1 null mice.Biochem Pharmacol. 2017 Jan 15;124:94-102. doi: 10.1016/j.bcp.2016.11.012. Epub 2016 Nov 11. Biochem Pharmacol. 2017. PMID: 27845049 Free PMC article.
-
Functional and Biochemical Consequences of Disease Variants in Neurotransmitter Transporters: A Special Emphasis on Folding and Trafficking Deficits.Pharmacol Ther. 2021 Jun;222:107785. doi: 10.1016/j.pharmthera.2020.107785. Epub 2020 Dec 10. Pharmacol Ther. 2021. PMID: 33310157 Free PMC article. Review.
-
In Vitro Fertilisation of Mouse Oocytes in L-Proline and L-Pipecolic Acid Improves Subsequent Development.Cells. 2021 May 29;10(6):1352. doi: 10.3390/cells10061352. Cells. 2021. PMID: 34072568 Free PMC article.
-
The Saccharomyces cerevisiae amino acid transporter Lyp1 has a broad substrate spectrum.FEBS Lett. 2025 Jun;599(11):1609-1621. doi: 10.1002/1873-3468.70044. Epub 2025 Apr 18. FEBS Lett. 2025. PMID: 40247776 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

