A hierarchical model for incomplete alignments in phylogenetic inference
- PMID: 19147663
- PMCID: PMC2647833
- DOI: 10.1093/bioinformatics/btp015
A hierarchical model for incomplete alignments in phylogenetic inference
Abstract
Motivation: Full-length DNA and protein sequences that span the entire length of a gene are ideally used for multiple sequence alignments (MSAs) and the subsequent inference of their relationships. Frequently, however, MSAs contain a substantial amount of missing data. For example, expressed sequence tags (ESTs), which are partial sequences of expressed genes, are the predominant source of sequence data for many organisms. The patterns of missing data typical for EST-derived alignments greatly compromise the accuracy of estimated phylogenies.
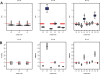
Results: We present a statistical method for inferring phylogenetic trees from EST-based incomplete MSA data. We propose a class of hierarchical models for modeling pairwise distances between the sequences, and develop a fully Bayesian approach for estimation of the model parameters. Once the distance matrix is estimated, the phylogenetic tree may be constructed by applying neighbor-joining (or any other algorithm of choice). We also show that maximizing the marginal likelihood from the Bayesian approach yields similar results to a profile likelihood estimation. The proposed methods are illustrated using simulated protein families, for which the true phylogeny is known, and one real protein family.
Availability: R code for fitting these models are available from: http://people.bu.edu/gupta/software.htm.
Figures

Similar articles
-
Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment?BMC Evol Biol. 2008 Mar 26;8:95. doi: 10.1186/1471-2148-8-95. BMC Evol Biol. 2008. PMID: 18366758 Free PMC article.
-
On the quality of tree-based protein classification.Bioinformatics. 2005 May 1;21(9):1876-90. doi: 10.1093/bioinformatics/bti244. Epub 2005 Jan 12. Bioinformatics. 2005. PMID: 15647305
-
Bayesian coestimation of phylogeny and sequence alignment.BMC Bioinformatics. 2005 Apr 1;6:83. doi: 10.1186/1471-2105-6-83. BMC Bioinformatics. 2005. PMID: 15804354 Free PMC article.
-
Genome BLAST distance phylogenies inferred from whole plastid and whole mitochondrion genome sequences.BMC Bioinformatics. 2006 Jul 19;7:350. doi: 10.1186/1471-2105-7-350. BMC Bioinformatics. 2006. PMID: 16854218 Free PMC article.
-
Revisiting Evaluation of Multiple Sequence Alignment Methods.Methods Mol Biol. 2021;2231:299-317. doi: 10.1007/978-1-0716-1036-7_17. Methods Mol Biol. 2021. PMID: 33289899 Review.
Cited by
-
PhyloMissForest: a random forest framework to construct phylogenetic trees with missing data.BMC Genomics. 2022 May 18;23(1):377. doi: 10.1186/s12864-022-08540-6. BMC Genomics. 2022. PMID: 35585494 Free PMC article.
-
Selecting informative subsets of sparse supermatrices increases the chance to find correct trees.BMC Bioinformatics. 2013 Dec 3;14:348. doi: 10.1186/1471-2105-14-348. BMC Bioinformatics. 2013. PMID: 24299043 Free PMC article.
References
-
- Anderson J. The phylogenetic trunk: maximal inclusion of taxa with missing data in an analysis of the lepospondyli. Syst. Biol. 2001;50:170–193. - PubMed
-
- Bevan R, et al. Accounting for gene rate heterogeneity in phylogenetic inference. Syst. Biol. 2007;56:194–205. - PubMed
-
- Bininda-Emonds OR. The evolution of supertrees. Trends Ecol. Evol. 2004;19:315–322. - PubMed
-
- Bouck A, Vision TJ. The molecular ecologist's guide to expressed sequence tags. Mol. Ecol. 2007;16:907–924. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

