Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group
- PMID: 19147825
- PMCID: PMC2630563
- DOI: 10.2353/ajpath.2009.080631
Molecular classification of rhabdomyosarcoma--genotypic and phenotypic determinants of diagnosis: a report from the Children's Oncology Group
Abstract
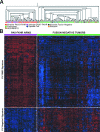
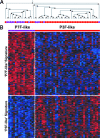
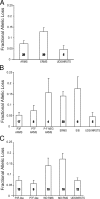
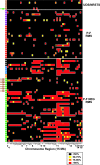
Rhabdomyosarcoma (RMS) in children occurs as two major histological subtypes, embryonal (ERMS) and alveolar (ARMS). ERMS is associated with an 11p15.5 loss of heterozygosity (LOH) and may be confused with nonmyogenic, non-RMS soft tissue sarcomas. ARMS expresses the product of a genomic translocation that fuses FOXO1 (FKHR) with either PAX3 or PAX7 (P-F); however, at least 25% of cases lack these translocations. Here, we describe a genomic-based classification scheme that is derived from the combined gene expression profiling and LOH analysis of 160 cases of RMS and non-RMS soft tissue sarcomas that is at variance with conventional histopathological schemes. We found that gene expression profiles and patterns of LOH of ARMS cases lacking P-F translocations are indistinguishable from conventional ERMS cases. A subset of tumors that has been histologically classified as RMS lack myogenic gene expression. However, classification based on gene expression is possible using as few as five genes with an estimated error rate of less than 5%. Using immunohistochemistry, we characterized two markers, HMGA2 and TFAP2ss, which facilitate the differential diagnoses of ERMS and P-F RMS, respectively, using clinical material. These objectively derived molecular classes are based solely on genomic analysis at the time of diagnosis and are highly reproducible. Adoption of these molecular criteria may offer a more clinically relevant diagnostic scheme, thus potentially improving patient management and therapeutic RMS outcomes.
Figures


References
-
- Asmar L, Gehan EA, Newton WA, Webber BL, Marsden HB, van Unnik AJ, Hamoudi AB, Shimada H, Tsokos M, Harms D. Agreement among and within groups of pathologists in the classification of rhabdomyosarcoma and related childhood sarcomas. Report of an international study of four pathology classifications. Cancer. 1994;74:2579–2588. - PubMed
-
- Enterline HT, Horn RC., Jr Alveolar rhabdomyosarcoma; a distinctive tumor type. Am J Clin Pathol. 1958;29:356–366. - PubMed
-
- Newton WA, Jr, Gehan EA, Webber BL, Marsden HB, van Unnik AJ, Hamoudi AB, Tsokos MG, Shimada H, Harms D, Schmidt D. Classification of rhabdomyosarcomas and related sarcomas. Pathologic aspects and proposal for a new classification—an Intergroup Rhabdomyosarcoma Study. Cancer. 1995;76:1073–1085. - PubMed
-
- Riopelle JL, Thériault JP. Sur une forme méconnue de sarcome des parties molles: le rhabdomyosarcome alvéolaire. Ann Anat Pathol (Paris) 1956;1:88–111. - PubMed
-
- Tsokos M, Webber BL, Parham DM, Wesley RA, Miser A, Miser JS, Etcubanas E, Kinsella T, Grayson J, Glatstein E. Rhabdomyosarcoma: a new classification scheme related to prognosis. Arch Pathol Lab Med. 1992;116:847–855. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

