DNA damage tolerance: when it's OK to make mistakes
- PMID: 19148176
- PMCID: PMC2663399
- DOI: 10.1038/nchembio.139
DNA damage tolerance: when it's OK to make mistakes
Abstract
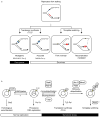
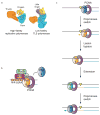
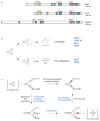
Mutations can be beneficial under conditions in which genetic diversity is advantageous, such as somatic hypermutation and antibody generation, but they can also be lethal when they disrupt basic cellular processes or cause uncontrolled proliferation and cancer. Mutations arise from inaccurate processing of lesions generated by endogenous and exogenous DNA damaging agents, and the genome is particularly vulnerable to such damage during S phase. In this phase of the cell cycle, many lesions in the DNA template block replication. Such lesions must be bypassed in order to preserve fork stability and to ensure completion of DNA replication. Lesion bypass is carried out by a set of error-prone and error-free processes collectively referred to as DNA damage tolerance mechanisms. Here, we discuss how two types of DNA damage tolerance, translesion synthesis and template switching, are regulated at stalled replication forks by ubiquitination of PCNA, and the conditions under which they occur.
Figures
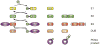

References
-
- Friedberg EC. Suffering in silence: the tolerance of DNA damage. Nat Rev Mol Cell Biol. 2005;6:943–953. - PubMed
-
- Nyberg KA, Michelson RJ, Putnam CW, Weinert TA. Toward maintaining the genome: DNA damage and replication checkpoints. Annu Rev Genet. 2002;36:617–656. - PubMed
-
- Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6:953–966. - PubMed
-
- Andersen PL, Xu F, Xiao W. Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 2008;18:162–173. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

