miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1
- PMID: 19148268
- PMCID: PMC2607557
- DOI: 10.1371/journal.ppat.1000263
miR-198 inhibits HIV-1 gene expression and replication in monocytes and its mechanism of action appears to involve repression of cyclin T1
Abstract
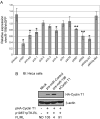
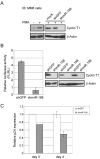
Cyclin T1 is a regulatory subunit of a general RNA polymerase II elongation factor known as P-TEFb. Cyclin T1 is also required for Tat transactivation of HIV-1 LTR-directed gene expression. Translation of Cyclin T1 mRNA has been shown to be repressed in human monocytes, and this repression is relieved when cells differentiate to macrophages. We identified miR-198 as a microRNA (miRNA) that is strongly down-regulated when monocytes are induced to differentiate. Ectopic expression of miR-198 in tissue culture cells reduced Cyclin T1 protein expression, and plasmid reporter assays verified miR-198 target sequences in the 3' untranslated region (3'UTR) of Cyclin T1 mRNA. Cyclin T1 protein levels increased when an inhibitor of miR-198 was transfected into primary monocytes, and overexpression of miR-198 in primary monocytes repressed the normal up-regulation of Cyclin T1 during differentiation. Expression of an HIV-1 proviral plasmid and HIV-1 replication were repressed in a monocytic cell line upon overexpression of miR-198. Our data indicate that miR-198 functions to restrict HIV-1 replication in monocytes, and its mechanism of action appears to involve repression of Cyclin T1 expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






Similar articles
-
Regulation of cyclin T1 and HIV-1 Replication by microRNAs in resting CD4+ T lymphocytes.J Virol. 2012 Mar;86(6):3244-52. doi: 10.1128/JVI.05065-11. Epub 2011 Dec 28. J Virol. 2012. PMID: 22205749 Free PMC article.
-
Induction of the HIV-1 Tat co-factor cyclin T1 during monocyte differentiation is required for the regulated expression of a large portion of cellular mRNAs.Retrovirology. 2006 Jun 9;3:32. doi: 10.1186/1742-4690-3-32. Retrovirology. 2006. PMID: 16764723 Free PMC article.
-
Interleukin-10 inhibits HIV-1 LTR-directed gene expression in human macrophages through the induction of cyclin T1 proteolysis.Virology. 2006 Sep 1;352(2):485-92. doi: 10.1016/j.virol.2006.05.013. Epub 2006 Jun 15. Virology. 2006. PMID: 16781761
-
Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages.Curr HIV Res. 2003 Oct;1(4):395-404. doi: 10.2174/1570162033485159. Curr HIV Res. 2003. PMID: 15049426 Review.
-
HIV-1 infection and regulation of Tat function in macrophages.Int J Biochem Cell Biol. 2004 Sep;36(9):1767-75. doi: 10.1016/j.biocel.2004.02.018. Int J Biochem Cell Biol. 2004. PMID: 15183343 Review.
Cited by
-
MicroRNA regulation and its effects on cellular transcriptome in human immunodeficiency virus-1 (HIV-1) infected individuals with distinct viral load and CD4 cell counts.BMC Infect Dis. 2013 May 30;13:250. doi: 10.1186/1471-2334-13-250. BMC Infect Dis. 2013. PMID: 23721325 Free PMC article.
-
Expression profiling of human milk derived exosomal microRNAs and their targets in HIV-1 infected mothers.Sci Rep. 2020 Jul 31;10(1):12931. doi: 10.1038/s41598-020-69799-x. Sci Rep. 2020. PMID: 32737406 Free PMC article. Clinical Trial.
-
Human immunodeficiency virus (HIV) latency: the major hurdle in HIV eradication.Mol Med. 2012 Sep 25;18(1):1096-108. doi: 10.2119/molmed.2012.00194. Mol Med. 2012. PMID: 22692576 Free PMC article. Review.
-
Identification and analysis of differential miRNAs in PK-15 cells after foot-and-mouth disease virus infection.PLoS One. 2014 Mar 27;9(3):e90865. doi: 10.1371/journal.pone.0090865. eCollection 2014. PLoS One. 2014. PMID: 24675746 Free PMC article.
-
HIV and the Macrophage: From Cell Reservoirs to Drug Delivery to Viral Eradication.J Neuroimmune Pharmacol. 2019 Mar;14(1):52-67. doi: 10.1007/s11481-018-9785-6. Epub 2018 Mar 23. J Neuroimmune Pharmacol. 2019. PMID: 29572681 Free PMC article. Review.
References
-
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. - PubMed
-
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem. 1999;274:34527–34530. - PubMed
-
- Rice AP, Herrmann CH. Regulation of TAK/P-TEFb in CD4+ T lymphocytes and macrophages. Curr HIV Res. 2003;1:395–404. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

