Deletion hotspots in AMACR promoter CpG island are cis-regulatory elements controlling the gene expression in the colon
- PMID: 19148275
- PMCID: PMC2613032
- DOI: 10.1371/journal.pgen.1000334
Deletion hotspots in AMACR promoter CpG island are cis-regulatory elements controlling the gene expression in the colon
Abstract
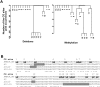
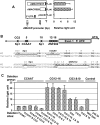
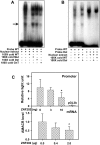
Alpha-methylacyl-coenzyme A racemase (AMACR) regulates peroxisomal beta-oxidation of phytol-derived, branched-chain fatty acids from red meat and dairy products -- suspected risk factors for colon carcinoma (CCa). AMACR was first found overexpressed in prostate cancer but not in benign glands and is now an established diagnostic marker for prostate cancer. Aberrant expression of AMACR was recently reported in Cca; however, little is known about how this gene is abnormally activated in cancer. By using a panel of immunostained-laser-capture-microdissected clinical samples comprising the entire colon adenoma-carcinoma sequence, we show that deregulation of AMACR during colon carcinogenesis involves two nonrandom events, resulting in the mutually exclusive existence of double-deletion at CG3 and CG10 and deletion of CG12-16 in a newly identified CpG island within the core promoter of AMACR. The double-deletion at CG3 and CG10 was found to be a somatic lesion. It existed in histologically normal colonic glands and tubular adenomas with low AMACR expression and was absent in villous adenomas and all CCas expressing variable levels of AMACR. In contrast, deletion of CG12-16 was shown to be a constitutional allele with a frequency of 43% in a general population. Its prevalence reached 89% in moderately differentiated CCas strongly expressing AMACR but only existed at 14% in poorly differentiated CCas expressing little or no AMACR. The DNA sequences housing these deletions were found to be putative cis-regulatory elements for Sp1 at CG3 and CG10, and ZNF202 at CG12-16. Chromatin immunoprecipitation, siRNA knockdown, gel shift assay, ectopic expression, and promoter analyses supported the regulation by Sp1 and ZNF202 of AMACR gene expression in an opposite manner. Our findings identified key in vivo events and novel transcription factors responsible for AMACR regulation in CCas and suggested these AMACR deletions may have diagnostic/prognostic value for colon carcinogenesis.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
A dietary enzyme: alpha-methylacyl-CoA racemase/P504S is overexpressed in colon carcinoma.Cancer Detect Prev. 2003;27(6):422-6. doi: 10.1016/j.cdp.2003.07.003. Cancer Detect Prev. 2003. PMID: 14642549
-
A nonclassic CCAAT enhancer element binding protein binding site contributes to alpha-methylacyl-CoA racemase expression in prostate cancer.Mol Cancer Res. 2005 Feb;3(2):110-8. doi: 10.1158/1541-7786.MCR-04-0178. Mol Cancer Res. 2005. PMID: 15755877
-
Molecular cloning and preliminary analysis of the human alpha-methylacyl-CoA racemase promoter.Mol Biol Rep. 2009 Mar;36(3):423-30. doi: 10.1007/s11033-007-9196-x. Epub 2007 Dec 14. Mol Biol Rep. 2009. PMID: 18080842
-
Discovery and clinical application of a novel prostate cancer marker: alpha-methylacyl CoA racemase (P504S).Am J Clin Pathol. 2004 Aug;122(2):275-89. doi: 10.1309/EJUY-UQPE-X1MG-68MK. Am J Clin Pathol. 2004. PMID: 15323145 Review.
-
Phytanic acid, AMACR and prostate cancer risk.Future Oncol. 2006 Apr;2(2):213-23. doi: 10.2217/14796694.2.2.213. Future Oncol. 2006. PMID: 16563090 Review.
Cited by
-
Mapping cancer biology in space: applications and perspectives on spatial omics for oncology.Mol Cancer. 2024 Jan 30;23(1):26. doi: 10.1186/s12943-024-01941-z. Mol Cancer. 2024. PMID: 38291400 Free PMC article. Review.
-
Increased expression of α-methylacyl-coenzyme A racemase (AMACR; p504s) and p16 in distal hyperplastic polyps.Diagn Pathol. 2013 Oct 23;8:178. doi: 10.1186/1746-1596-8-178. Diagn Pathol. 2013. PMID: 24152881 Free PMC article.
-
Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration.Nat Commun. 2018 Feb 16;9(1):700. doi: 10.1038/s41467-018-03019-z. Nat Commun. 2018. PMID: 29453456 Free PMC article.
-
Methylation of a single intronic CpG mediates expression silencing of the PMP24 gene in prostate cancer.Prostate. 2010 May 15;70(7):765-76. doi: 10.1002/pros.21109. Prostate. 2010. PMID: 20054818 Free PMC article.
-
Electrical Stimulation Enhances Cardiac Differentiation of Human Induced Pluripotent Stem Cells for Myocardial Infarction Therapy.Antioxid Redox Signal. 2018 Feb 10;28(5):371-384. doi: 10.1089/ars.2016.6766. Epub 2017 Jan 12. Antioxid Redox Signal. 2018. PMID: 27903111 Free PMC article.
References
-
- Ferdinandusse S, Rusch H, van Lint AE, Dacremont G, Wanders RJ, et al. Stereochemistry of the peroxisomal branched-chain fatty acid alpha- and beta-oxidation systems in patients suffering from different peroxisomal disorders. J Lipid Res. 2002;43:438–444. - PubMed
-
- Baron A, Migita T, Tang D, Loda M. Fatty acid synthase: a metabolic oncogene in prostate cancer? J Cell Biochem. 2004;91:47–53. - PubMed
-
- Zha S, Ferdinandusse S, Hicks JL, Denis S, Dunn TA, et al. Peroxisomal branched chain fatty acid beta-oxidation pathway is upregulated in prostate cancer. Prostate. 2005;63:316–323. - PubMed
-
- Carnell AJ, Hale I, Denis S, Wanders RJ, Isaacs WB, et al. Design, synthesis, and in vitro testing of alpha-methylacyl-CoA racemase inhibitors. J Med Chem. 2007;50:2700–2707. - PubMed
-
- Zha S, Isaacs WB. A nonclassic CCAAT enhancer element binding protein binding site contributes to alpha-methylacyl-CoA racemase expression in prostate cancer. Mol Cancer Res. 2005;3:110–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

