Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics
- PMID: 19150357
- PMCID: PMC2644216
- DOI: 10.1016/j.jmb.2008.12.064
Recombineering reveals a diverse collection of ribosomal proteins L4 and L22 that confer resistance to macrolide antibiotics
Abstract
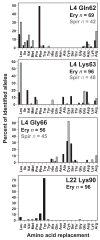
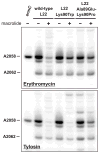
Mutations in ribosomal proteins L4 and L22 confer resistance to erythromycin and other macrolide antibiotics in a variety of bacteria. L4 and L22 have elongated loops whose tips converge in the peptide exit tunnel near the macrolide-binding site, and resistance mutations typically affect residues within these loops. Here, we used bacteriophage lambda Red-mediated recombination, or "recombineering," to uncover new L4 and L22 alleles that confer macrolide resistance in Escherichia coli. We randomized residues at the tips of the L4 and L22 loops using recombineered oligonucleotide libraries and selected the mutagenized cells for erythromycin-resistant mutants. These experiments led to the identification of 341 resistance mutations encoding 278 unique L4 and L22 proteins-the overwhelming majority of which are novel. Many resistance mutations were complex, involving multiple missense mutations, in-frame deletions, and insertions. Transfer of L4 and L22 mutations into wild-type cells by phage P1-mediated transduction demonstrated that each allele was sufficient to confer macrolide resistance. Although L4 and L22 mutants are typically resistant to most macrolides, selections carried out on different antibiotics revealed macrolide-specific resistance mutations. L22 Lys90Trp is one such allele that confers resistance to erythromycin but not to tylosin and spiramycin. Purified L22 Lys90Trp ribosomes show reduced erythromycin binding but have the same affinity for tylosin as wild-type ribosomes. Moreover, dimethyl sulfate methylation protection assays demonstrated that L22 Lys90Trp ribosomes bind tylosin more readily than erythromycin in vivo. This work underscores the exceptional functional plasticity of the L4 and L22 proteins and highlights the utility of Red-mediated recombination in targeted genetic selections.
Figures

References
-
- Hansen JL, Ippolito JA, Ban N, Nissen P, Moore PB, Steitz TA. The structures of four macrolide antibiotics bound to the large ribosomal subunit. Mol Cell. 2002;10:117–128. - PubMed
-
- Schlunzen F, Harms JM, Franceschi F, Hansen HA, Bartels H, Zarivach R, Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. - PubMed
-
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. - PubMed
-
- Tenson T, Lovmar M, Ehrenberg M. The mechanism of action of macrolides, lincosamides and streptogramin B reveals the nascent peptide exit path in the ribosome. J Mol Biol. 2003;330:1005–1014. - PubMed
-
- Champney WS. Bacterial ribosomal subunit synthesis: a novel antibiotic target. Curr Drug Targets Infect Disord. 2001;1:19–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

