New information content in RNA base pairing deduced from quantitative analysis of high-resolution structures
- PMID: 19150407
- PMCID: PMC2681097
- DOI: 10.1016/j.ymeth.2008.12.003
New information content in RNA base pairing deduced from quantitative analysis of high-resolution structures
Abstract
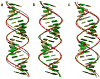
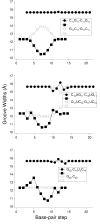
Non-canonical base pairs play important roles in organizing the complex three-dimensional folding of RNA. Here, we outline methodology developed both to analyze the spatial patterns of interacting base pairs in known RNA structures and to reconstruct models from the collective experimental information. We focus attention on the structural context and deformability of the seven pairing patterns found in greatest abundance in the helical segments in a set of well-resolved crystal structures, including (i-ii) the canonical A.U and G.C Watson-Crick base pairs, (iii) the G.U wobble pair, (iv) the sheared G.A pair, (v) the A.U Hoogsteen pair, (vi) the U.U wobble pair, and (vii) the G.A Watson-Crick-like pair. The non-canonical pairs stand out from the canonical associations in terms of apparent deformability, spanning a broader range of conformational states as measured by the six rigid-body parameters used to describe the spatial arrangements of the interacting bases, the root-mean-square deviations of the base-pair atoms, and the fluctuations in hydrogen-bonding geometry. The deformabilties, the modes of base-pair deformation, and the preferred sites of occurrence depend on sequence. We also characterize the positioning and overlap of the base pairs with respect to the base pairs that stack immediately above and below them in double-helical fragments. We incorporate the observed positions of the bases, base pairs, and intervening phosphorus atoms in models to predict the effects of the non-canonical interactions on overall helical structure.
Figures



Similar articles
-
Stacking geometry for non-canonical G:U wobble base pair containing dinucleotide sequences in RNA: dispersion-corrected DFT-D study.Biopolymers. 2015 Jun;103(6):328-38. doi: 10.1002/bip.22616. Biopolymers. 2015. PMID: 25652776
-
Effects of Noncanonical Base Pairing on RNA Folding: Structural Context and Spatial Arrangements of G·A Pairs.Biochemistry. 2019 May 21;58(20):2474-2487. doi: 10.1021/acs.biochem.9b00122. Epub 2019 May 8. Biochemistry. 2019. PMID: 31008589 Free PMC article.
-
Diversity of base-pair conformations and their occurrence in rRNA structure and RNA structural motifs.J Mol Biol. 2004 Dec 10;344(5):1225-49. doi: 10.1016/j.jmb.2004.09.072. J Mol Biol. 2004. PMID: 15561141
-
An innate twist between Crick's wobble and Watson-Crick base pairs.RNA. 2013 Aug;19(8):1038-53. doi: 10.1261/rna.036905.112. RNA. 2013. PMID: 23861536 Free PMC article. Review.
-
RNA structure and dynamics: a base pairing perspective.Prog Biophys Mol Biol. 2013 Nov;113(2):264-83. doi: 10.1016/j.pbiomolbio.2013.07.003. Epub 2013 Jul 23. Prog Biophys Mol Biol. 2013. PMID: 23891726 Review.
Cited by
-
Unusual target site disruption by the rare-cutting HNH restriction endonuclease PacI.Structure. 2010 Jun 9;18(6):734-43. doi: 10.1016/j.str.2010.03.009. Structure. 2010. PMID: 20541511 Free PMC article.
-
Revisiting DNA Sequence-Dependent Deformability in High-Resolution Structures: Effects of Flanking Base Pairs on Dinucleotide Morphology and Global Chain Configuration.Life (Basel). 2022 May 20;12(5):759. doi: 10.3390/life12050759. Life (Basel). 2022. PMID: 35629425 Free PMC article.
-
RNABPDB: Molecular Modeling of RNA Structure-From Base Pair Analysis in Crystals to Structure Prediction.Interdiscip Sci. 2022 Sep;14(3):759-774. doi: 10.1007/s12539-022-00528-w. Epub 2022 Jun 15. Interdiscip Sci. 2022. PMID: 35705797
-
Site-directed mutants of 16S rRNA reveal important RNA domains for KsgA function and 30S subunit assembly.Biochemistry. 2011 Feb 8;50(5):854-63. doi: 10.1021/bi101005r. Epub 2011 Jan 11. Biochemistry. 2011. PMID: 21142019 Free PMC article.
-
RNA structural motifs that entail hydrogen bonds involving sugar-phosphate backbone atoms of RNA.New J Chem. 2010 May 1;34(5):910-917. doi: 10.1039/b9nj00754g. New J Chem. 2010. PMID: 20689681 Free PMC article.
References
-
- Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. - PubMed
-
- Mazière P, Enright AJ. Prediction of microRNA targets. Drug Discov Today. 2007;12:452–458. - PubMed
-
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

