Structure-guided Mutational Analysis of the Nucleotidyltransferase Domain of Escherichia coli DNA Ligase (LigA)
- PMID: 19150981
- PMCID: PMC2659207
- DOI: 10.1074/jbc.M808476200
Structure-guided Mutational Analysis of the Nucleotidyltransferase Domain of Escherichia coli DNA Ligase (LigA)
Abstract
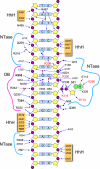
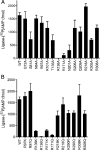
NAD(+)-dependent DNA ligases (LigA) are ubiquitous in bacteria, where they are essential for growth and present attractive targets for antimicrobial drug discovery. LigA has a distinctive modular structure in which a nucleotidyltransferase catalytic domain is flanked by an upstream NMN-binding module and by downstream OB-fold, zinc finger, helix-hairpin-helix, and BRCT domains. Here we conducted a structure-function analysis of the nucleotidyltransferase domain of Escherichia coli LigA, guided by the crystal structure of the LigA-DNA-adenylate intermediate. We tested the effects of 29 alanine and conservative mutations at 15 amino acids on ligase activity in vitro and in vivo. We thereby identified essential functional groups that coordinate the reactive phosphates (Arg(136)), contact the AMP adenine (Lys(290)), engage the phosphodiester backbone flanking the nick (Arg(218), Arg(308), Arg(97) plus Arg(101)), or stabilize the active domain fold (Arg(171)). Finer analysis of the mutational effects revealed step-specific functions for Arg(136), which is essential for the reaction of LigA with NAD(+) to form the covalent ligase-AMP intermediate (step 1) and for the transfer of AMP to the nick 5'-PO(4) to form the DNA-adenylate intermediate (step 2) but is dispensable for phosphodiester formation at a preadenylylated nick (step 3).
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

