Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system
- PMID: 19151083
- PMCID: PMC2655697
- DOI: 10.1093/nar/gkp001
Unconventional decoding of the AUA codon as methionine by mitochondrial tRNAMet with the anticodon f5CAU as revealed with a mitochondrial in vitro translation system
Abstract
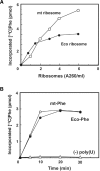
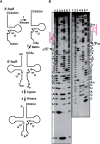
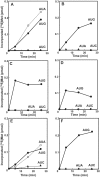
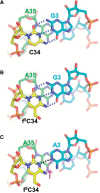
Mitochondrial (mt) tRNA(Met) has the unusual modified nucleotide 5-formylcytidine (f(5)C) in the first position of the anticodon. This tRNA must translate both AUG and AUA as methionine. By constructing an in vitro translation system from bovine liver mitochondria, we examined the decoding properties of the native mt tRNA(Met) carrying f(5)C in the anticodon compared to a transcript that lacks the modification. The native mt Met-tRNA could recognize both AUA and AUG codons as Met, but the corresponding synthetic tRNA(Met) lacking f(5)C (anticodon CAU), recognized only the AUG codon in both the codon-dependent ribosomal binding and in vitro translation assays. Furthermore, the Escherichia coli elongator tRNA(Met)(m) with the anticodon ac(4)CAU (ac(4)C = 4-acetylcytidine) and the bovine cytoplasmic initiator tRNA(Met) (anticodon CAU) translated only the AUG codon for Met on mt ribosome. The codon recognition patterns of these tRNAs were the same on E. coli ribosomes. These results demonstrate that the f(5)C modification in mt tRNA(Met) plays a crucial role in decoding the nonuniversal AUA codon as Met, and that the genetic code variation is compensated by a change in the tRNA anticodon, not by a change in the ribosome. Base pairing models of f(5)C-G and f(5)C-A based on the chemical properties of f(5)C are presented.
Figures


References
-
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. - PubMed
-
- Himeno H, Masaki H, Kawai T, Ohta T, Kumagai I, Miura K, Watanabe K. Unusual genetic codes and a novel gene structure for tRNA(AGYSer) in starfish mitochondrial DNA. Gene. 1987;56:219–230. - PubMed
-
- Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. Int. Rev. Cytol. 1992;141:173–216. - PubMed
-
- Xu X, Arnason U. The complete mitochondrial DNA sequence of the horse, Equus caballus: extensive heteroplasmy of the control region. Gene. 1994;148:357–362. - PubMed
-
- Krettek A, Gullberg A, Arnason U. Sequence analysis of the complete mitochondrial DNA molecule of the hedgehog, Ernaceus europaeus, and the phylogenic position of the Lipotyphla. J. Mol. Evol. 1995;41:952–957. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

