Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis
- PMID: 19151092
- PMCID: PMC2655667
- DOI: 10.1093/nar/gkn1039
Aminoacyl-tRNA recognition by the FemXWv transferase for bacterial cell wall synthesis
Abstract
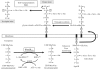
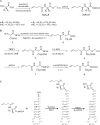
Transferases of the Fem family catalyse peptide-bond formation by using aminoacyl-tRNAs and peptidoglycan precursors as donor and acceptor substrates, respectively. The specificity of Fem transferases is essential since mis-incorporated amino acids could act as chain terminators thereby preventing formation of a functional stress-bearing peptidoglycan network. Here we have developed chemical acylation of RNA helices with natural and non-proteinogenic amino acids to gain insight into the specificity of the model transferase FemX(Wv). Combining modifications in the RNA and aminoacyl moieties of the donor substrate revealed that unfavourable interactions of FemX(Wv) with the acceptor arm of tRNA(Gly) and with L-Ser or larger residues quantitatively accounts for the preferential transfer of L-Ala observed with complete aminoacyl-tRNAs. The main FemX(Wv) identity determinant was identified as the penultimate base pair (G(2)-C(71)) of the acceptor arm instead of G(3)*U(70) for the alanyl-tRNA synthetase. FemX(Wv) tolerated a configuration inversion of the Calpha of L-Ala but not the introduction of a second methyl on this atom. These results indicate that aminoacyl-tRNA recognition by FemX(Wv) is distinct from other components of the translation machinery and relies on the exclusion of bulky amino acids and of the sequence of tRNA(Gly) from the active site.
Figures






References
-
- den Blaauwen T, de Pedro MA, Nguyen-Distèche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol. Rev. 2008;32:321–344. - PubMed
-
- Dramsi S, Magnet S, Davison S, Arthur M. Covalent attachment of proteins to peptidoglycan. FEMS Microbiol. Rev. 2008;32:307–320. - PubMed
-
- Vollmer W. Structural variation in the glycan strands of bacterial peptidoglycan. FEMS Microbiol. Rev. 2008;32:287–306. - PubMed
-
- Kern T, Hediger S, Muller P, Giustini C, Joris B, Bougault C, Vollmer W, Simorre JP. Toward the characterization of peptidoglycan structure and protein-peptidoglycan interactions by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2008;130:5618–5619. - PubMed
-
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 2008;32:149–167. - PubMed

