Correlation between biofilm formation and the hypoxic response in Candida parapsilosis
- PMID: 19151323
- PMCID: PMC2669199
- DOI: 10.1128/EC.00350-08
Correlation between biofilm formation and the hypoxic response in Candida parapsilosis
Abstract
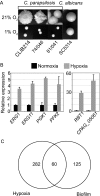
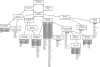
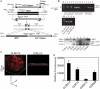
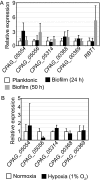
The ability of Candida parapsilosis to form biofilms on indwelling medical devices is correlated with virulence. To identify genes that are important for biofilm formation, we used arrays representing approximately 4,000 open reading frames (ORFs) to compare the transcriptional profile of biofilm cells growing in a microfermentor under continuous flow conditions with that of cells in planktonic culture. The expression of genes involved in fatty acid and ergosterol metabolism and in glycolysis, is upregulated in biofilms. The transcriptional profile of C. parapsilosis biofilm cells resembles that of Candida albicans cells grown under hypoxic conditions. We therefore subsequently used whole-genome arrays (representing 5,900 ORFs) to determine the hypoxic response of C. parapsilosis and showed that the levels of expression of genes involved in the ergosterol and glycolytic pathways, together with several cell wall genes, are increased. Our results indicate that there is substantial overlap between the hypoxic responses of C. parapsilosis and C. albicans and that this may be important for biofilm development. Knocking out an ortholog of the cell wall gene RBT1, whose expression is induced both in biofilms and under conditions of hypoxia in C. parapsilosis, reduces biofilm development.
Figures
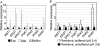
References
-
- Alexa, A., J. Rahnenfuhrer, and T. Lengauer. 2006. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 221600-1607. - PubMed
-
- Almirante, B., D. Rodriguez, M. Cuenca-Estrella, M. Almela, F. Sanchez, J. Ayats, C. Alonso-Tarres, J. L. Rodriguez-Tudela, and A. Pahissa. 2006. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 441681-1685. - PMC - PubMed
-
- An, D., and M. R. Parsek. 2007. The promise and peril of transcriptional profiling in biofilm communities. Curr. Opin. Microbiol. 10292-296. - PubMed
-
- Baillie, G. S., and L. J. Douglas. 1999. Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48671-679. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

