Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples
- PMID: 19151326
- PMCID: PMC2653237
- DOI: 10.1128/EC.00215-08
Uneven distribution of mating types among genotypes of Candida glabrata isolates from clinical samples
Abstract
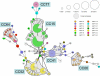
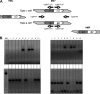
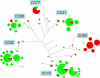
In order to shed light on its basic biology, we initiated a population genetic analysis of Candida glabrata, an emerging pathogenic yeast with no sexual stage yet recognized. A worldwide collection of clinical strains was subjected to analysis using variable number of tandem repeats (VNTR) at nine loci. The clustering of strains obtained with this method was congruent with that obtained using sequence polymorphism of the NMT1 gene, a locus previously proposed for lineage assignment. Linkage disequilibrium supported the hypothesis of a mainly clonal reproduction. No heterozygous diploid genotype was found. Minimum-spanning tree analysis of VNTR data revealed clonal expansions and associated genotypic diversification. Mating type analysis revealed that 80% of the strains examined are MATa and 20% MATalpha and that the two alleles are not evenly distributed. The MATa genotype dominated within large clonal groups that contained only one or a few MATalpha types. In contrast, two groups were dominated by MATalpha strains. Our data are consistent with rare independent mating type switching events occurring preferentially from type a to alpha, although the alternative possibility of selection favoring type a isolates cannot be excluded.
Figures
References
-
- Agapow, P. M., and A. Burt. 2001. Indices of multilocus linkage disequilibrium. Mol. Ecol. Notes 1101-102.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

