Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells
- PMID: 19151716
- PMCID: PMC2632725
- DOI: 10.1038/ng.297
Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells
Abstract
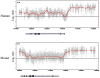
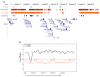
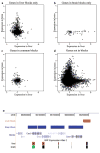
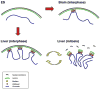
Higher eukaryotes must adapt a totipotent genome to specialized cell types with stable but limited functions. One potential mechanism for lineage restriction is changes in chromatin, and differentiation-related chromatin changes have been observed for individual genes. We have taken a genome-wide view of histone H3 lysine 9 dimethylation (H3K9Me2) and find that differentiated tissues show surprisingly large K9-modified regions (up to 4.9 Mb). These regions are highly conserved between human and mouse and are differentiation specific, covering only approximately 4% of the genome in undifferentiated mouse embryonic stem (ES) cells, compared to 31% in differentiated ES cells, approximately 46% in liver and approximately 10% in brain. These modifications require histone methyltransferase G9a and are inversely related to expression of genes within the regions. We term these regions large organized chromatin K9 modifications (LOCKs). LOCKs are substantially lost in cancer cell lines, and they may provide a cell type-heritable mechanism for phenotypic plasticity in development and disease.
Figures

Comment in
-
Reassessing the abundance of H3K9me2 chromatin domains in embryonic stem cells.Nat Genet. 2010 Jan;42(1):4; author reply 5-6. doi: 10.1038/ng0110-4. Nat Genet. 2010. PMID: 20037608 No abstract available.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

