Conformational dynamics of the flexible catalytic loop in Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase
- PMID: 19152632
- PMCID: PMC2982673
- DOI: 10.1111/j.1747-0285.2008.00749.x
Conformational dynamics of the flexible catalytic loop in Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase
Abstract
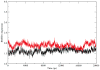
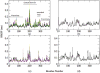
In mycobacteria, the biosynthesis of the precursors to the essential isoprenoids, isopentenyl diphosphate and dimethylallyl pyrophosphate is carried out by the methylerythritol phosphate pathway. This route of synthesis is absent in humans, who utilize the alternative mevalonate acid route, thus making the enzymes of the methylerythritol phosphate pathway of chemotherapeutic interest. One such identified target is the second enzyme of the pathway, 1-deoxy-D-xylulose 5-phosphate reductoisomerase. Only limited information is currently available concerning the catalytic mechanism and structural dynamics of this enzyme, and only recently has a crystal structure of Mycobacterium tuberculosis species of this enzyme been resolved including all factors required for binding. Here, the dynamics of the enzyme is studied in complex with NADPH, Mn2+, in the presence and absence of the fosmidomycin inhibitor using conventional molecular dynamics and an enhanced sampling technique, reversible digitally filtered molecular dynamics. The simulations reveal significant differences in the conformational dynamics of the vital catalytic loop between the inhibitor-free and inhibitor-bound enzyme complexes and highlight the contributions of conserved residues in this region. The substantial fluctuations observed suggest that 1-deoxy-D-xylulose 5-phosphate reductoisomerase may be a promising target for computer-aided drug discovery through the relaxed complex method.
Figures




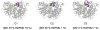

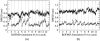
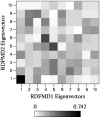
Similar articles
-
The 1.9 A resolution structure of Mycobacterium tuberculosis 1-deoxy-D-xylulose 5-phosphate reductoisomerase, a potential drug target.Acta Crystallogr D Biol Crystallogr. 2006 Jul;62(Pt 7):807-13. doi: 10.1107/S0907444906019196. Epub 2006 Jun 20. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16790937
-
Structures of Mycobacterium tuberculosis 1-deoxy-D-xylulose-5-phosphate reductoisomerase provide new insights into catalysis.J Biol Chem. 2007 Jul 6;282(27):19905-16. doi: 10.1074/jbc.M701935200. Epub 2007 May 9. J Biol Chem. 2007. PMID: 17491006
-
Mutation in the flexible loop of 1-deoxy-D-xylulose 5-phosphate reductoisomerase broadens substrate utilization.Arch Biochem Biophys. 2005 Dec 15;444(2):159-64. doi: 10.1016/j.abb.2005.10.004. Epub 2005 Oct 27. Arch Biochem Biophys. 2005. PMID: 16289362
-
1-Deoxy-D-xylulose 5-phosphate reductoisomerase: an overview.Bioorg Chem. 2004 Dec;32(6):483-93. doi: 10.1016/j.bioorg.2004.08.004. Bioorg Chem. 2004. PMID: 15530989 Review.
-
Targeting the methyl erythritol phosphate (MEP) pathway for novel antimalarial, antibacterial and herbicidal drug discovery: inhibition of 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR) enzyme.Curr Pharm Des. 2007;13(11):1161-77. doi: 10.2174/138161207780618939. Curr Pharm Des. 2007. PMID: 17430177 Review.
Cited by
-
Tryptophan as a molecular shovel in the glycosyl transfer activity of Trypanosoma cruzi trans-sialidase.Biophys J. 2010 May 19;98(9):L38-40. doi: 10.1016/j.bpj.2010.01.006. Biophys J. 2010. PMID: 20441732 Free PMC article.
-
Deconstructing honeybee vitellogenin: novel 40 kDa fragment assigned to its N terminus.J Exp Biol. 2011 Feb 15;214(Pt 4):582-92. doi: 10.1242/jeb.048314. J Exp Biol. 2011. PMID: 21270306 Free PMC article.
-
A computational study of the molecular basis of antibiotic resistance in a DXR mutant.J Comput Aided Mol Des. 2019 Oct;33(10):927-940. doi: 10.1007/s10822-019-00229-5. Epub 2019 Oct 26. J Comput Aided Mol Des. 2019. PMID: 31654265
-
From Zn to Mn: the study of novel manganese-binding groups in the search for new drugs against tuberculosis.Chem Biol Drug Des. 2011 Feb;77(2):117-23. doi: 10.1111/j.1747-0285.2010.01060.x. Chem Biol Drug Des. 2011. PMID: 21266015 Free PMC article.
-
Inhibition of the Fe(4)S(4)-cluster-containing protein IspH (LytB): electron paramagnetic resonance, metallacycles, and mechanisms.J Am Chem Soc. 2010 May 19;132(19):6719-27. doi: 10.1021/ja909664j. J Am Chem Soc. 2010. PMID: 20426416 Free PMC article.
References
-
- WHO. Global tuberculosis control: surveillance, planning, financing: WHO report 2008. World Health Organization; 2008. Global tuberculosis control: surveillance, planning, financing: WHO report 2008.
-
- Ebrahim GJ. Drug resistance in tuberculosis. J Trop Pediatr. 2007;53:147–9. - PubMed
-
- Sacchettini JC, Poulter CD. Biochemistry - Creating isoprenoid diversity. Science. 1997;277:1788–9. - PubMed
-
- Swanson KM, Hohl RJ. Anti-cancer therapy: Targeting the mevalonate pathway. Curr Cancer Drug Targets. 2006;6:15–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

