Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer
- PMID: 19153141
- PMCID: PMC2655672
- DOI: 10.1093/nar/gkp002
Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer
Abstract
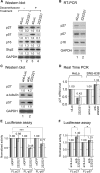
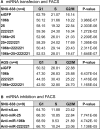
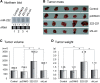
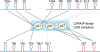
microRNAs (miRNAs) play integral roles in diverse processes including tumorigenesis. miRNA gene loci are often found in close conjunction, and such clustered miRNA genes are transcribed from a common promoter to generate polycistronic primary transcript. The primary transcript (pri-miRNA) is then processed by two RNase III proteins to release the mature miRNAs. Although it has been speculated that the miRNAs in the same cluster may play related biological functions, this has not been experimentally addressed. Here we report that the miRNAs in two clusters (miR-106b approximately 93 approximately 25 and miR-222 approximately 221) suppress the Cip/Kip family members of Cdk inhibitors (p57(Kip2), p21(Cip1) and p27(Kip1)). We show that miR-25 targets p57 through the 3'-UTR. Furthermore, miR-106b and miR-93 control p21 while miR-222 and miR-221 regulate both p27 and p57. Ectopic expression of these miRNAs results in activation of Cdk2 and facilitation of G1/S phase transition. Consistent with these results, both clusters are abnormally upregulated in gastric cancer tissues compared to the corresponding normal tissues. Ectopic expression of miR-222 cluster enhanced tumor growth in the mouse xenograft model. Our study demonstrates the functional associations between clustered miRNAs and further implicates that effective cancer treatment may require a combinatorial approach to target multiple oncogenic miRNA clusters.
Figures



References
-
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 2008;9:102–114. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

