Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation
- PMID: 19153223
- PMCID: PMC2654299
- DOI: 10.1083/jcb.200806072
Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation
Abstract
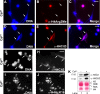
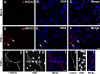
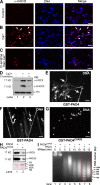
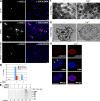
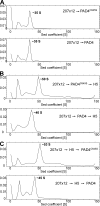
Peripheral blood neutrophils form highly decondensed chromatin structures, termed neutrophil extracellular traps (NETs), that have been implicated in innate immune response to bacterial infection. Neutrophils express high levels of peptidylarginine deiminase 4 (PAD4), which catalyzes histone citrullination. However, whether PAD4 or histone citrullination plays a role in chromatin structure in neutrophils is unclear. In this study, we show that the hypercitrullination of histones by PAD4 mediates chromatin decondensation. Histone hypercitrullination is detected on highly decondensed chromatin in HL-60 granulocytes and blood neutrophils. The inhibition of PAD4 decreases histone hypercitrullination and the formation of NET-like structures, whereas PAD4 treatment of HL-60 cells facilitates these processes. The loss of heterochromatin and multilobular nuclear structures is detected in HL-60 granulocytes after PAD4 activation. Importantly, citrullination of biochemically defined avian nucleosome arrays inhibits their compaction by the linker histone H5 to form higher order chromatin structures. Together, these results suggest that histone hypercitrullination has important functions in chromatin decondensation in granulocytes/neutrophils.
Figures
References
-
- Beiter K., Wartha F., Albiger B., Normark S., Zychlinsky A., Henriques-Normark B. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps.Curr. Biol. 16:401–407 - PubMed
-
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. 2004. Neutrophil extracellular traps kill bacteria.Science. 303:1532–1535 - PubMed
-
- Buchanan J.T., Simpson A.J., Aziz R.K., Liu G.Y., Kristian S.A., Kotb M., Feramisco J., Nizet V. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps.Curr. Biol. 16:396–400 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

