The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase
- PMID: 19153239
- PMCID: PMC2655585
- DOI: 10.1128/JVI.01849-08
The NS4A protein of hepatitis C virus promotes RNA-coupled ATP hydrolysis by the NS3 helicase
Abstract
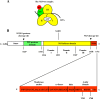
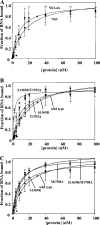
Nonstructural protein 3 (NS3) is an essential replicative component of the hepatitis C virus (HCV) and a member of the DExH/D-box family of proteins. The C-terminal region of NS3 (NS3hel) exhibits RNA-stimulated NTPase and helicase activity, while the N-terminal serine protease domain of NS3 enhances RNA binding and unwinding by NS3hel. The nonstructural protein 4A (NS4A) binds to the NS3 protease domain and serves as an obligate cofactor for NS3 serine protease activity. Given its role in stimulating protease activity, we sought to determine whether NS4A also influences the activity of NS3hel. Here we show that NS4A enhances the ability of NS3hel to bind RNA in the presence of ATP, thereby acting as a cofactor for helicase activity. This effect is mediated by amino acids in the C-terminal acidic domain of NS4A. When these residues are mutated, one observes drastic reductions in ATP-coupled RNA binding and duplex unwinding by NS3. These same mutations are lethal in HCV replicons, thereby establishing in vitro and in vivo that NS4A plays an important role in the helicase mechanism of NS3 and its function in replication.
Figures



References
-
- Appel, N., T. Schaller, F. Penin, and R. Bartenschlager. 2006. From structure to function: new insights into hepatitis C virus RNA replication. J. Biol. Chem. 2819833-9836. - PubMed
-
- Beran, R. K., M. M. Bruno, H. A. Bowers, E. Jankowsky, and A. M. Pyle. 2006. Robust translocation along a molecular monorail: the NS3 helicase from hepatitis C virus traverses unusually large disruptions in its track. J. Mol. Biol. 358974-982. - PubMed
-
- Beran, R. K., V. Serebrov, and A. M. Pyle. 2007. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J. Biol. Chem. 28234913-34920. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

