Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3
- PMID: 19153604
- PMCID: PMC2646149
- DOI: 10.1038/emboj.2008.289
Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3
Abstract
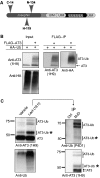
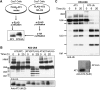
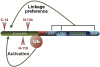
Deubiquitinating enzymes (DUBs) control the ubiquitination status of proteins in various cellular pathways. Regulation of the activity of DUBs, which is critically important to cellular homoeostasis, can be achieved at the level of gene expression, protein complex formation, or degradation. Here, we report that ubiquitination also directly regulates the activity of a DUB, ataxin-3, a polyglutamine disease protein implicated in protein quality control pathways. Ubiquitination enhances ubiquitin (Ub) chain cleavage by ataxin-3, but does not alter its preference for K63-linked Ub chains. In cells, ubiquitination of endogenous ataxin-3 increases when the proteasome is inhibited, when excess Ub is present, or when the unfolded protein response is induced, suggesting that the cellular functions of ataxin-3 in protein quality control are modulated through ubiquitination. Ataxin-3 is the first reported DUB in which ubiquitination directly regulates catalytic activity. We propose a new function for protein ubiquitination in regulating the activity of certain DUBs and perhaps other enzymes.
Figures





Similar articles
-
Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117.J Biol Chem. 2010 Dec 10;285(50):39303-13. doi: 10.1074/jbc.M110.181610. Epub 2010 Oct 13. J Biol Chem. 2010. PMID: 20943656 Free PMC article.
-
Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity.J Biol Chem. 2007 Oct 5;282(40):29348-58. doi: 10.1074/jbc.M704126200. Epub 2007 Aug 10. J Biol Chem. 2007. PMID: 17693639
-
The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability.Hum Mol Genet. 2011 Jan 1;20(1):141-54. doi: 10.1093/hmg/ddq452. Epub 2010 Oct 11. Hum Mol Genet. 2011. PMID: 20940148 Free PMC article.
-
The role of deubiquitinating enzymes in apoptosis.Cell Mol Life Sci. 2011 Jan;68(1):15-26. doi: 10.1007/s00018-010-0504-6. Epub 2010 Aug 21. Cell Mol Life Sci. 2011. PMID: 20730552 Free PMC article. Review.
-
Toward therapeutic targets for SCA3: Insight into the role of Machado-Joseph disease protein ataxin-3 in misfolded proteins clearance.Prog Neurobiol. 2015 Sep;132:34-58. doi: 10.1016/j.pneurobio.2015.06.004. Epub 2015 Jun 27. Prog Neurobiol. 2015. PMID: 26123252 Review.
Cited by
-
Physiological and pathophysiological characteristics of ataxin-3 isoforms.J Biol Chem. 2019 Jan 11;294(2):644-661. doi: 10.1074/jbc.RA118.005801. Epub 2018 Nov 19. J Biol Chem. 2019. PMID: 30455355 Free PMC article.
-
Linear poly-ubiquitin remodels the proteome and influences hundreds of regulators in Drosophila.G3 (Bethesda). 2024 Nov 6;14(11):jkae209. doi: 10.1093/g3journal/jkae209. G3 (Bethesda). 2024. PMID: 39325835 Free PMC article.
-
Allosteric Modulation of Pathological Ataxin-3 Aggregation: A Path to Spinocerebellar Ataxia Type-3 Therapies.bioRxiv [Preprint]. 2025 Jan 24:2025.01.22.633970. doi: 10.1101/2025.01.22.633970. bioRxiv. 2025. PMID: 39896516 Free PMC article. Preprint.
-
Autophagy induction reduces mutant ataxin-3 levels and toxicity in a mouse model of spinocerebellar ataxia type 3.Brain. 2010 Jan;133(Pt 1):93-104. doi: 10.1093/brain/awp292. Epub 2009 Dec 9. Brain. 2010. PMID: 20007218 Free PMC article.
-
Roles of p97-associated deubiquitinases in protein quality control at the endoplasmic reticulum.Curr Protein Pept Sci. 2012 Aug;13(5):436-46. doi: 10.2174/138920312802430608. Curr Protein Pept Sci. 2012. PMID: 22812527 Free PMC article. Review.
References
-
- Amerik AY, Hochstrasser M (2004) Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta 1695: 189–207 - PubMed
-
- Bowman AB, Lam YC, Jafar-Nejad P, Chen HK, Richman R, Samaco RC, Fryer JD, Kahle JJ, Orr HT, Zoghbi HY (2007) Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet 39: 373–379 - PubMed
-
- Brzovic PS, Lissounov A, Christensen DE, Hoyt DW, Klevit RE (2006) A UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol Cell 21: 873–880 - PubMed
-
- Burnett B, Li F, Pittman RN (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet 12: 3195–3205 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

