Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals
- PMID: 19153606
- PMCID: PMC2646147
- DOI: 10.1038/emboj.2008.281
Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals
Erratum in
- EMBO J. 2009 Apr 8;28(7):992
Abstract
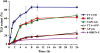
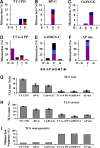
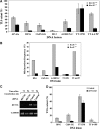
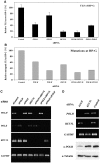
DNA replication across blocking lesions occurs by translesion DNA synthesis (TLS), involving a multitude of mutagenic DNA polymerases that operate to protect the mammalian genome. Using a quantitative TLS assay, we identified three main classes of TLS in human cells: two rapid and error-free, and the third slow and error-prone. A single gene, REV3L, encoding the catalytic subunit of DNA polymerase zeta (pol zeta), was found to have a pivotal role in TLS, being involved in TLS across all lesions examined, except for a TT cyclobutane dimer. Genetic epistasis siRNA analysis indicated that discrete two-polymerase combinations with pol zeta dictate error-prone or error-free TLS across the same lesion. These results highlight the central role of pol zeta in both error-prone and error-free TLS in mammalian cells, and show that bypass of a single lesion may involve at least three different DNA polymerases, operating in different two-polymerase combinations.
Figures


Comment in
-
Bypass specialists operate together.EMBO J. 2009 Feb 18;28(4):313-4. doi: 10.1038/emboj.2008.303. EMBO J. 2009. PMID: 19225445 Free PMC article. Review.
References
-
- Adar S, Livneh Z (2006) Translesion DNA synthesis across non-DNA segments in cultured human cells. DNA Repair (Amst) 5: 479–490 - PubMed
-
- Alt A, Lammens K, Chiocchini C, Lammens A, Pieck JC, Kuch D, Hopfner KP, Carell T (2007) Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta. Science 318: 967–970 - PubMed
-
- Avkin S, Goldsmith M, Velasco-Miguel S, Geacintov N, Friedberg EC, Livneh Z (2004) Quantitative analysis of translesion DNA synthesis across a benzo[a]pyrene-guanine adduct in mammalian cells. The role of DNA polymerase κ. J Biol Chem 279: 53298–53305 - PubMed
-
- Avkin S, Sevilya Z, Toube L, Geacintov NE, Chaney SG, Oren M, Livneh Z (2006) p53 and p21 regulate error-prone DNA repair to yield a lower mutation load. Mol Cell 22: 407–413 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

