Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry
- PMID: 19154110
- PMCID: PMC2681235
- DOI: 10.1021/ac8019202
Peptide quantification using 8-plex isobaric tags and electron transfer dissociation tandem mass spectrometry
Abstract
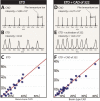
Isobaric tags for absolute and relative quantitation (iTRAQ) allow for simultaneous relative quantification of peptides from up to eight different samples. Typically peptides labeled with 8-plex iTRAQ tags are pooled and fragmented using beam-type collision activated dissociation (CAD) which, in addition to cleaving the peptide backbone bonds, cleaves the tag to produce reporter ions. The relative intensities of the reporters are directly proportional to the relative abundances of each peptide in the solution phase. Recently, studies using the 4-plex iTRAQ tagging reagent demonstrated that electron transfer dissociation (ETD) of 4-plex iTRAQ labeled peptides cleaves at the N-C alpha bond in the tag and allows for up to three channels of quantification. In this paper we investigate the ETD fragmentation patterns of peptides labeled with 8-plex iTRAQ tags. We demonstrate that upon ETD, peptides labeled with 8-plex iTRAQ tags fragment to produce unique reporter ions that allow for five channels of quantification. ETD-MS/MS of these labeled peptides also produces a peak at 322 m/z which, upon resonant excitation (CAD), gives rise to all eight iTRAQ reporter ions and allows for eight channels of quantification. Comparison of this method to beam-type CAD quantification shows a good correlation (y = 0.91x + 0.01, R(2) = 0.9383).
Figures



References
-
- Aebersold R, Mann M. Nature. 2003;422:198–207. - PubMed
-
- Bantscheff M, Schirle M, Sweetman G, Rick J, Kuster B. Anal. Bioanal. Chem. 2007;389:1017–1031. - PubMed
-
- Conrads TP, Issaq HJ, Veenstra TD. Biochem. Biophys. Res. Commun. 2002;290:885–890. - PubMed
-
- Heck AJR, Krijgsveld J. Expert Rev. Proteomics. 2004;1:317–326. - PubMed
-
- Julka S, Regnier F. J. Proteome Res. 2004;3:350–363. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

