Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component
- PMID: 19155210
- PMCID: PMC2722196
- DOI: 10.1093/hmg/ddp036
Deficiency of the E3 ubiquitin ligase TRIM32 in mice leads to a myopathy with a neurogenic component
Abstract
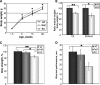
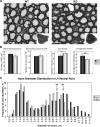
Limb-girdle muscular dystrophy type 2H (LGMD2H) and sarcotubular myopathy are hereditary skeletal muscle disorders caused by mutations in TRIM32. We previously identified TRIM32 as an E3 ubiquitin ligase that binds to myosin and ubiquitinates actin. To date four TRIM32 mutations have been linked to LGMD2H, all of which occur in the C-terminal NHL domains. Unexpectedly, a fifth mutation in the B-box of TRIM32 causes a completely different, multisystemic disorder, Bardet-Biedl syndrome type 11. It is not understood how allelic mutations in TRIM32 can create such diverse phenotypic outcomes. To generate a tool for elucidating the complex in vivo functions of TRIM32, we created the first murine Trim32 knock-out model (T32KO). Histological analysis of T32KO skeletal muscles revealed mild myopathic changes. Electron microscopy showed areas with Z-line streaming and a dilated sarcotubular system with vacuoles -- the latter being a prominent feature of sarcotubular myopathy. Therefore, our model replicates phenotypes of LGMD2H and sarcotubular myopathy. The level of Trim32 expression in normal mouse brain exceeds that observed in skeletal muscle by more than 100 times, as we demonstrated by real-time PCR. Intriguingly, analysis of T32KO neural tissue revealed a decreased concentration of neurofilaments and a reduction in myelinated motoraxon diameters. The axonal changes suggest a shift toward a slower motor unit type. Not surprisingly, T32KO soleus muscle expressed an elevated type I slow myosin isotype with a concomitant reduction in the type II fast myosin. These data suggest that muscular dystrophy due to TRIM32 mutations involves both neurogenic and myogenic characteristics.
Figures






References
-
- Kudryashova E., Kudryashov D., Kramerova I., Spencer M.J. Trim32 is a ubiquitin ligase mutated in limb girdle muscular dystrophy type 2H that binds to skeletal muscle myosin and ubiquitinates actin. J. Mol. Biol. 2005;354:413–424. - PubMed
-
- Albor A., El-Hizawi S., Horn E.J., Laederich M., Frosk P., Wrogemann K., Kulesz-Martin M. The interaction of Piasy with Trim32, an E3-ubiquitin ligase mutated in limb-girdle muscular dystrophy type 2H, promotes Piasy degradation and regulates UVB-induced keratinocyte apoptosis through NFκB. J. Biol. Chem. 2006;281:25850–25866. - PubMed
-
- Kano S., Miyajima N., Fukuda S., Hatakeyama S. Tripartite motif protein 32 facilitates cell growth and migration via degradation of Abl-interactor 2. Cancer Res. 2008;68:5572–5580. - PubMed
-
- Pickart C.M., Eddins M.J. Ubiquitin: structures, functions, mechanisms. Biochim. Biophys. Acta. 2004;1695:55–72. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

