The Electron Transfer Pathway of the Na+-pumping NADH:Quinone Oxidoreductase from Vibrio cholerae
- PMID: 19155212
- PMCID: PMC2659253
- DOI: 10.1074/jbc.M809395200
The Electron Transfer Pathway of the Na+-pumping NADH:Quinone Oxidoreductase from Vibrio cholerae
Abstract
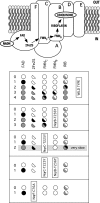
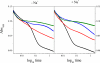
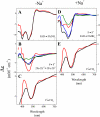
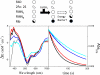
The Na(+)-pumping NADH:quinone oxidoreductase (Na(+)-NQR) is the only respiratory enzyme that operates as a Na(+) pump. This redox-driven Na(+) pump is amenable to experimental approaches not available for H(+) pumps, providing an excellent system for mechanistic studies of ion translocation. An understanding of the internal electron transfer steps and their Na(+) dependence is an essential prerequisite for such studies. To this end, we analyzed the reduction kinetics of the wild type Na(+)-NQR, as well as site-directed mutants of the enzyme, which lack specific cofactors. NADH and ubiquinol were used as reductants in separate experiments, and a full spectrum UV-visible stopped flow kinetic method was employed. The results make it possible to define the complete sequence of redox carriers in the electrons transfer pathway through the enzyme. Electrons flow from NADH to quinone through the FAD in subunit F, the 2Fe-2S center, the FMN in subunit C, the FMN in subunit B, and finally riboflavin. The reduction of the FMN(C) to its anionic flavosemiquinone state is the first Na(+)-dependent process, suggesting that reduction of this site is linked to Na(+) uptake. During the reduction reaction, two FMNs are transformed to their anionic flavosemiquinone in a single kinetic step. Subsequently, FMN(C) is converted to the flavohydroquinone, accounting for the single anionic flavosemiquinone radical in the fully reduced enzyme. A model of the electron transfer steps in the catalytic cycle of Na(+)-NQR is presented to account for the kinetic and spectroscopic data.
Figures




References
-
- Barquera, B., Hellwig, P., Zhou, W., Morgan, J. E., Hase, C. C., Gosink, K. K., Nilges, M., Bruesehoff, P. J., Roth, A., Lancaster, C. R., and Gennis, R. B. (2002) Biochemistry 41 3781-3789 - PubMed
-
- Dibrov, P. A., Kostryko, V. A., Lazarova, R. L., Skulachev, V. P., and Smirnova, I. A. (1986) Biochim. Biophys. Acta 850 449-457 - PubMed
-
- Bogachev, A. V., and Verkhovsky, M. I. (2005) Biochemistry (Mosc.) 70 143-149 - PubMed
-
- Bogachev, A. V., Murtazina, R. A., and Skulachev, V. P. (1997) FEBS Lett. 409 475-477 - PubMed
-
- Stolpe, S., and Friedrich, T. (2004) J. Biol. Chem. 279 18377-18383 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

