Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae
- PMID: 19155276
- PMCID: PMC2655692
- DOI: 10.1093/nar/gkp003
Histone H3 Lys79 methylation is required for efficient nucleotide excision repair in a silenced locus of Saccharomyces cerevisiae
Abstract
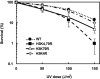
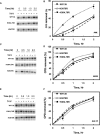
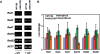
Methylation of specific histone lysine residues regulates gene expression and heterochromatin function, but little is known about its role in DNA repair. To examine how changes in conserved methylated residues of histone H3 affect nucleotide excision repair (NER), viable H3K4R and H3K79R mutants were generated in Saccharomyces cerevisiae. These mutants show decreased UV survival and impaired NER at the transcriptionally silent HML locus, while maintaining normal NER in the constitutively expressed RPB2 gene and transcriptionally repressed, nucleosome loaded GAL10 gene. Moreover, the HML chromatin in these mutants has reduced accessibility to Micrococcal nuclease (MNase). Importantly, chromatin immunoprecipitation analysis demonstrates there is enhanced recruitment of the Sir complex at the HML locus of these mutants, and deletion of the SIR2 or SIR3 genes restores the MNase accessibility and DNA repair efficiency at this locus. Furthermore, following UV irradiation expression of NER genes in these mutants remains at wild type levels, with the exception of RAD16 which decreases by more than 2-fold. These results indicate that impaired NER occurs in the silenced chromatin of H3K79R and H3K4,79R mutants as a result of increased binding of Sir complexes, which may reduce DNA lesion accessibility to repair enzymes.
Figures



Similar articles
-
Silenced yeast chromatin is maintained by Sir2 in preference to permitting histone acetylations for efficient NER.Nucleic Acids Res. 2010 Aug;38(14):4675-86. doi: 10.1093/nar/gkq242. Epub 2010 Apr 12. Nucleic Acids Res. 2010. PMID: 20385597 Free PMC article.
-
Regulated acetylation and deacetylation of H4 K16 is essential for efficient NER in Saccharomyces cerevisiae.DNA Repair (Amst). 2018 Dec;72:39-55. doi: 10.1016/j.dnarep.2018.09.009. Epub 2018 Sep 22. DNA Repair (Amst). 2018. PMID: 30274769
-
A single amino acid change in histone H4 enhances UV survival and DNA repair in yeast.Nucleic Acids Res. 2008 Jun;36(11):3857-66. doi: 10.1093/nar/gkn311. Epub 2008 May 28. Nucleic Acids Res. 2008. PMID: 18508805 Free PMC article.
-
The Sir proteins of Saccharomyces cerevisiae: mediators of transcriptional silencing and much more.Curr Opin Microbiol. 2000 Apr;3(2):132-7. doi: 10.1016/s1369-5274(00)00064-3. Curr Opin Microbiol. 2000. PMID: 10744999 Review.
-
The Nuts and Bolts of Transcriptionally Silent Chromatin in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1563-99. doi: 10.1534/genetics.112.145243. Genetics. 2016. PMID: 27516616 Free PMC article. Review.
Cited by
-
Loss of H3 K79 trimethylation leads to suppression of Rtt107-dependent DNA damage sensitivity through the translesion synthesis pathway.J Biol Chem. 2010 Nov 5;285(45):35113-22. doi: 10.1074/jbc.M110.116855. Epub 2010 Sep 1. J Biol Chem. 2010. PMID: 20810656 Free PMC article.
-
NuA4 acetyltransferase is required for efficient nucleotide excision repair in yeast.DNA Repair (Amst). 2019 Jan;73:91-98. doi: 10.1016/j.dnarep.2018.11.006. Epub 2018 Nov 14. DNA Repair (Amst). 2019. PMID: 30473425 Free PMC article.
-
Regulation of UV damage repair in quiescent yeast cells.DNA Repair (Amst). 2020 Jun;90:102861. doi: 10.1016/j.dnarep.2020.102861. Epub 2020 Apr 30. DNA Repair (Amst). 2020. PMID: 32403026 Free PMC article.
-
Histone ubiquitylation and its roles in transcription and DNA damage response.DNA Repair (Amst). 2015 Dec;36:36-42. doi: 10.1016/j.dnarep.2015.09.016. Epub 2015 Sep 16. DNA Repair (Amst). 2015. PMID: 26422137 Free PMC article. Review.
-
Photoreactivation Activities of Rad5, Rad16A and Rad16B Help Beauveria bassiana to Recover from Solar Ultraviolet Damage.J Fungi (Basel). 2024 Jun 13;10(6):420. doi: 10.3390/jof10060420. J Fungi (Basel). 2024. PMID: 38921406 Free PMC article.
References
-
- Luger K. Structure and dynamic behavior of nucleosomes. Curr. Opin. Genet. Dev. 2003;13:127–135. - PubMed
-
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. - PubMed
-
- Moazed D. Common themes in mechanisms of gene silencing. Mol. Cell. 2001;8:489–498. - PubMed
-
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. - PubMed
-
- Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

