The epigenetic profile of Ig genes is dynamically regulated during B cell differentiation and is modulated by pre-B cell receptor signaling
- PMID: 19155482
- PMCID: PMC2804480
- DOI: 10.4049/jimmunol.182.3.1362
The epigenetic profile of Ig genes is dynamically regulated during B cell differentiation and is modulated by pre-B cell receptor signaling
Abstract
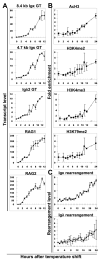
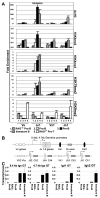
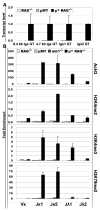
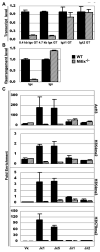
Ag receptor loci poised for V(D)J rearrangement undergo germline transcription (GT) of unrearranged genes, and the accessible gene segments are associated with posttranslational modifications (PTM) on histones. In this study, we performed a comprehensive analysis of the dynamic changes of four PTM throughout B and T cell differentiation in freshly isolated ex vivo cells. Methylation of lysines 4 and 79 of histone H3, and acetylation of H3, demonstrated stage and lineage specificity, and were most pronounced at the J segments of loci poised for, or undergoing, rearrangement, except for dimethylation of H3K4, which was more equally distributed on V, D, and J genes. Focusing on the IgL loci, we demonstrated there are no active PTM in the absence of pre-BCR signaling. The kappa locus GT and PTM on Jkappa genes are rapidly induced following pre-BCR signaling in large pre-B cells. In contrast, the lambda locus shows greatly delayed onset of GT and PTM, which do not reach high levels until the immature B cell compartment, the stage at which receptor editing is initiated. Analysis of MiEkappa(-/-) mice shows that this enhancer plays a key role in inducing not only GT, but PTM. Using an inducible pre-B cell line, we demonstrate that active PTM on Jkappa genes occur after GT is initiated, indicating that histone PTM do not make the Jkappa region accessible, but conversely, GT may play a role in adding PTM. Our data indicate that the epigenetic profile of IgL genes is dramatically modulated by pre-BCR signaling and B cell differentiation status.
Figures


References
-
- Martensson IL, Keenan RA, Licence S. The pre-B-cell receptor. Curr Opin Immunol. 2007;19:137–142. - PubMed
-
- Durdik J, Moore MW, Selsing E. Novel kappa light-chain gene rearrangements in mouse lambda light chain-producing B lymphocytes. Nature. 1984;307:749–752. - PubMed
-
- Engel H, Rolink A, Weiss S. B cells are programmed to activate kappa and lambda for rearrangement at consecutive developmental stages. Eur J Immunol. 1999;29:2167–2176. - PubMed
-
- Yancopoulos GD, Alt FW. Regulation of the assembly and expression of variable-region genes. Annu Rev Immunol. 1986;4:339–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

