Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer
- PMID: 19156145
- PMCID: PMC2658541
- DOI: 10.1038/sj.bjc.6604884
Dysregulation of the transcription factors SOX4, CBFB and SMARCC1 correlates with outcome of colorectal cancer
Abstract
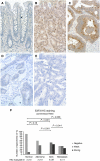
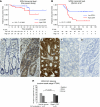
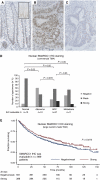
The aim of this study was to identify deregulated transcription factors (TFs) in colorectal cancer (CRC) and to evaluate their relation with the recurrence of stage II CRC and overall survival. Microarray-based transcript profiles of 20 normal mucosas and 424 CRC samples were used to identify 51 TFs displaying differential transcript levels between normal mucosa and CRC. For a subset of these we provide in vitro evidence that deregulation of the Wnt signalling pathway can lead to the alterations observed in tissues. Furthermore, in two independent cohorts of microsatellite-stable stage II cancers we found that high SOX4 transcript levels correlated with recurrence (HR 2.7; 95% CI, 1.2-6.0; P=0.01). Analyses of approximately 1000 stage I-III adenocarcinomas, by immunohistochemistry, revealed that patients with tumours displaying high levels of CBFB and SMARCC1 proteins had a significantly better overall survival rate (P=0.0001 and P=0.0275, respectively) than patients with low levels. Multivariate analyses revealed that a high CBFB protein level was an independent predictor of survival. In conclusion, several of the identified TFs seem to be involved in the progression of CRC.
Figures

References
-
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T (2006) SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res 66: 3434–3442 - PubMed
-
- Andersen CL, Jensen JL, Orntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64: 5245–5250 - PubMed
-
- Barrier A, Boelle PY, Roser F, Gregg J, Tse C, Brault D, Lacaine F, Houry S, Huguier M, Franc B, Flahault A, Lemoine A, Dudoit S (2006) Stage II colon cancer prognosis prediction by tumor gene expression profiling. J Clin Oncol 24: 4685–4691 - PubMed
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434: 864–870 - PubMed
-
- Birkenkamp-Demtroder K, Christensen LL, Olesen SH, Frederiksen CM, Laiho P, Aaltonen LA, Laurberg S, Sorensen FB, Hagemann R, Ørntoft TF (2002) Gene expression in colorectal cancer. Cancer Res 62: 4352–4363 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

