Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach
- PMID: 19156205
- PMCID: PMC2625441
- DOI: 10.1371/journal.pone.0004219
Direct metagenomic detection of viral pathogens in nasal and fecal specimens using an unbiased high-throughput sequencing approach
Abstract
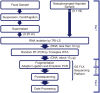
With the severe acute respiratory syndrome epidemic of 2003 and renewed attention on avian influenza viral pandemics, new surveillance systems are needed for the earlier detection of emerging infectious diseases. We applied a "next-generation" parallel sequencing platform for viral detection in nasopharyngeal and fecal samples collected during seasonal influenza virus (Flu) infections and norovirus outbreaks from 2005 to 2007 in Osaka, Japan. Random RT-PCR was performed to amplify RNA extracted from 0.1-0.25 ml of nasopharyngeal aspirates (N = 3) and fecal specimens (N = 5), and more than 10 microg of cDNA was synthesized. Unbiased high-throughput sequencing of these 8 samples yielded 15,298-32,335 (average 24,738) reads in a single 7.5 h run. In nasopharyngeal samples, although whole genome analysis was not available because the majority (>90%) of reads were host genome-derived, 20-460 Flu-reads were detected, which was sufficient for subtype identification. In fecal samples, bacteria and host cells were removed by centrifugation, resulting in gain of 484-15,260 reads of norovirus sequence (78-98% of the whole genome was covered), except for one specimen that was under-detectable by RT-PCR. These results suggest that our unbiased high-throughput sequencing approach is useful for directly detecting pathogenic viruses without advance genetic information. Although its cost and technological availability make it unlikely that this system will very soon be the diagnostic standard worldwide, this system could be useful for the earlier discovery of novel emerging viruses and bioterrorism, which are difficult to detect with conventional procedures.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

