Lupus-prone MRL/faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: concurrent upregulation of somatic hypermutation and class switch DNA recombination
- PMID: 19156553
- PMCID: PMC3140875
- DOI: 10.1080/08916930802629554
Lupus-prone MRL/faslpr/lpr mice display increased AID expression and extensive DNA lesions, comprising deletions and insertions, in the immunoglobulin locus: concurrent upregulation of somatic hypermutation and class switch DNA recombination
Abstract
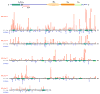
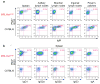
Systemic lupus erythematosus (SLE) is an autoimmune disease characterized by the production of an array of pathogenic autoantibodies, including high-affinity anti-dsDNA IgG antibodies. These autoantibodies are mutated and class-switched, mainly to IgG, indicating that immunoglobulin (Ig) gene somatic hypermutation (SHM) and class switch DNA recombination (CSR) are important in their generation. Lupus-prone MRL/fas(lpr/lpr) mice develop a systemic autoimmune syndrome that shares many features with human SLE. We found that Ig genes were heavily mutated in MRL/fas(lpr/lpr) mice and contained long stretches of DNA deletions and insertions. The spectrum of mutations in MRL/fas(lpr/lpr) B cells was significantly altered, including increased dG/dC transitions, increased targeting of the RGYW/WRCY mutational hotspot and the WGCW AID-targeting hotspot. We also showed that MRL/fas(lpr/lpr) greatly upregulated CSR, particularly to IgG2a and IgA in B cells of the spleen, lymph nodes and Peyer's patches. In MRL/fas(lpr/lpr) mice, the significant upregulation of SHM and CSR was associated with increased expression of activation-induced cytidine deaminase (AID), which mediates DNA lesion, the first step in SHM and CSR, and translesion DNA synthesis (TLS) polymerase (pol) theta, pol eta and pol zeta, which are involved in DNA synthesis/repair process associated with SHM and, possibly, CSR. Thus, in lupus-prone MRL/fas(lpr/lpr) mice, SHM and CSR are upregulated, as a result of enhanced AID expression and, therefore, DNA lesions, and dysregulated DNA repair factors, including TLS polymerases, which are involved in the repair process of AID-mediated DNA lesions.
Figures
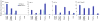




References
-
- Shlomchik MJ, Madaio MP. The role of antibodies and B cells in the pathogenesis of lupus nephritis. Springer Semin Immunopathol. 2003;24:363–375. - PubMed
-
- Diamond B. Autoimmunity. Immunol Rev. 2005;204:5–8. - PubMed
-
- van Es JH, Gmelig Meyling FH, van de Akker WR, Aanstoot H, Derksen RH, Logtenberg T. Somatic mutations in the variable regions of a human IgG anti-double-stranded DNA autoantibody suggest a role for antigen in the induction of systemic lupus erythematosus. J Exp Med. 1991;173:461–470. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous
