Production of random DNA oligomers for scalable DNA computing
- PMID: 19156734
- PMCID: PMC4991938
- DOI: 10.1002/biot.200800224
Production of random DNA oligomers for scalable DNA computing
Erratum in
- Biotechnol J. 2014 Nov;9(11):1458. Austin, Alan [corrected to Austin, Allen]
Abstract
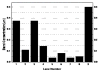
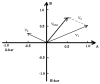
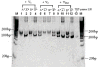
While remarkably complex networks of connected DNA molecules can form from a relatively small number of distinct oligomer strands, a large computational space created by DNA reactions would ultimately require the use of many distinct DNA strands. The automatic synthesis of this many distinct strands is economically prohibitive. We present here a new approach to producing distinct DNA oligomers based on the polymerase chain reaction (PCR) amplification of a few random template sequences. As an example, we designed a DNA template sequence consisting of a 50-mer random DNA segment flanked by two 20-mer invariant primer sequences. Amplification of a dilute sample containing about 30 different template molecules allows us to obtain around 10(11) copies of these molecules and their complements. We demonstrate the use of these amplicons to implement some of the vector operations that will be required in a DNA implementation of an analog neural network.
Figures



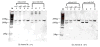



References
-
- Adleman LM. Molecular computation of solutions to combinatorial problems. Science. 1994;266:1021–1021. - PubMed
-
- Braich RS, Chelyapov N, Johnson C, Rothemund PWK, et al. Solution of a 20-Variable 3-SAT Problem on a DNA Computer. Science. 2002;296:499. - PubMed
-
- Winfree E, Liu FR, Wenzler LA, Seeman NC. Design and self-assembly of two-dimensional DNA crystals. Nature. 1998;394:539–544. - PubMed
-
- Seeman NC, Wang H, Yang XP, Liu FR, et al. New motifs in DNA nanotechnology. Nanotechnology. 1998;9:257–273.
-
- Liu QH, Wang LM, Frutos AG, Condon AE, et al. DNA computing on surfaces. Nature. 2000;403:175–179. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

