Distinct genomic aberrations associated with ERG rearranged prostate cancer
- PMID: 19156837
- PMCID: PMC2674964
- DOI: 10.1002/gcc.20647
Distinct genomic aberrations associated with ERG rearranged prostate cancer
Abstract
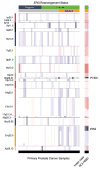
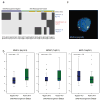
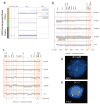
Emerging molecular and clinical data suggest that ETS fusion prostate cancer represents a distinct molecular subclass, driven most commonly by a hormonally regulated promoter and characterized by an aggressive natural history. The study of the genomic landscape of prostate cancer in the light of ETS fusion events is required to understand the foundation of this molecularly and clinically distinct subtype. We performed genome-wide profiling of 49 primary prostate cancers and identified 20 recurrent chromosomal copy number aberrations, mainly occurring as genomic losses. Co-occurring events included losses at 19q13.32 and 1p22.1. We discovered three genomic events associated with ERG rearranged prostate cancer, affecting 6q, 7q, and 16q. 6q loss in nonrearranged prostate cancer is accompanied by gene expression deregulation in an independent dataset and by protein deregulation of MYO6. To analyze copy number alterations within the ETS genes, we performed a comprehensive analysis of all 27 ETS genes and of the 3 Mbp genomic area between ERG and TMPRSS2 (21q) with an unprecedented resolution (30 bp). We demonstrate that high-resolution tiling arrays can be used to pin-point breakpoints leading to fusion events. This study provides further support to define a distinct molecular subtype of prostate cancer based on the presence of ETS gene rearrangements.
Figures


References
-
- Alers JC, Krijtenburg PJ, Vis AN, Hoedemaeker RF, Wildhagen MF, Hop WC, van Der Kwast TT, Schroder FH, Tanke HJ, van Dekken H. Molecular cytogenetic analysis of prostatic adenocarcinomas from screening studies: early cancers may contain aggressive genetic features. Am J Pathol. 2001;158:399–406. - PMC - PubMed
-
- Amin MB, Grignon DJ, Humphrey PA, Srigley JR. Gleason Grading of Prostate Cancer: a contemporary approach. 1. Philadelphia: Lippincott Williams and Wilkins; 2003. p. 116.
-
- Attard G, Clark J, Ambroisine L, Fisher G, Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, Moller H, Reuter V, De Bono JS, Scardino P, Cuzick J, Cooper CS. Duplication of the fusion of TMPRSS2 to ERG sequences identifies fatal human prostate cancer. Oncogene. 2008;27:253–263. - PMC - PubMed
-
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, Du J, Kau T, Thomas RK, Shah K, Soto H, Perner S, Prensner J, Debiasi RM, Demichelis F, Hatton C, Rubin MA, Garraway LA, Nelson SF, Liau L, Mischel PS, Cloughesy TF, Meyerson M, Golub TA, Lander ES, Mellinghoff IK, Sellers WR. Assessing the significance of chromosomal aberrations in cancer: Methodology and application to glioma. Proc Natl Acad Sci U S A. 2007;104:20007–20012. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

