Linking folding and binding
- PMID: 19157855
- PMCID: PMC2675572
- DOI: 10.1016/j.sbi.2008.12.003
Linking folding and binding
Abstract
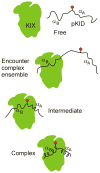
Many cellular proteins are intrinsically disordered and undergo folding, in whole or in part, upon binding to their physiological targets. The past few years have seen an exponential increase in papers describing characterization of intrinsically disordered proteins, both free and bound to targets. Although NMR spectroscopy remains the favored tool, a number of new biophysical techniques are proving exceptionally useful in defining the limits of the conformational ensembles. Advances have been made in prediction of the recognition elements in disordered proteins, in elucidating the kinetics and mechanism of the coupled folding and binding process, and in understanding the role of post-translational modifications in tuning the biological response. Here we review these and other recent advances that are providing new insights into the conformational propensities and interactions of intrinsically disordered proteins and are beginning to reveal general principles underlying their biological functions.
Figures
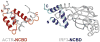
References
-
- Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. - PubMed
-
- Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets. The roles of intrinsic disorder in protein interaction networks. FEBS J. 2005;272:5129–5148. - PubMed
-
- Vucetic S, Xie HB, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6:1899–1916. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

