Self-assembly of Epstein-Barr virus capsids
- PMID: 19158247
- PMCID: PMC2663254
- DOI: 10.1128/JVI.01733-08
Self-assembly of Epstein-Barr virus capsids
Abstract
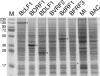
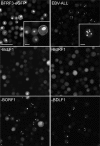
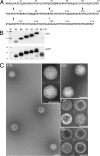
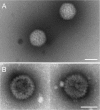
Epstein-Barr virus (EBV), a member of the Gammaherpesvirus family, primarily infects B lymphocytes and is responsible for a number of lymphoproliferative diseases. The molecular genetics of the assembly pathway and high-resolution structural analysis of the capsid have not been determined for this lymphocryptovirus. As a first step in studying EBV capsid assembly, the baculovirus expression vector (BEV) system was used to express the capsid shell proteins BcLF1 (major capsid protein), BORF1 (triplex protein), BDLF1 (triplex protein), and BFRF3 (small capsid protein); the internal scaffold protein, BdRF1; and the maturational protease (BVRF2). Coinfection of insect cells with the six viruses expressing these proteins resulted in the production of closed capsid structures as judged by electron microscopy and sedimentation methods. Therefore, as shown for other herpesviruses, only six proteins are required for EBV capsid assembly. Furthermore, the small capsid protein of EBV (BFRF3), like that of Kaposi's sarcoma-associated herpesvirus, was found to be required for assembly of a stable structure. Localization of the small capsid protein to nuclear assembly sites required both the major capsid (BcLF1) and scaffold proteins (BdRF1) but not the triplex proteins. Mutational analysis of BFRF3 showed that the N-terminal half (amino acids 1 to 88) of this polypeptide is required and sufficient for capsid assembly. A region spanning amino acids 65 to 88 is required for the concentration of BFRF3 at a subnuclear site and the N-terminal 65 amino acids contain the sequences required for interaction with major capsid protein. These studies have identified the multifunctional role of the gammaherpesvirus small capsid proteins.
Figures





Similar articles
-
Assembly of Epstein-Barr Virus Capsid in Promyelocytic Leukemia Nuclear Bodies.J Virol. 2015 Sep;89(17):8922-31. doi: 10.1128/JVI.01114-15. Epub 2015 Jun 17. J Virol. 2015. PMID: 26085145 Free PMC article.
-
Molecular interactions of Epstein-Barr virus capsid proteins.J Virol. 2011 Feb;85(4):1615-24. doi: 10.1128/JVI.01565-10. Epub 2010 Dec 8. J Virol. 2011. PMID: 21147928 Free PMC article.
-
Small capsid protein pORF65 is essential for assembly of Kaposi's sarcoma-associated herpesvirus capsids.J Virol. 2008 Jul;82(14):7201-11. doi: 10.1128/JVI.00423-08. Epub 2008 May 7. J Virol. 2008. PMID: 18463150 Free PMC article.
-
Herpesvirus capsid assembly: insights from structural analysis.Curr Opin Virol. 2011 Aug;1(2):142-9. doi: 10.1016/j.coviro.2011.06.003. Curr Opin Virol. 2011. PMID: 21927635 Free PMC article. Review.
-
The Ins and Outs of Herpesviral Capsids: Divergent Structures and Assembly Mechanisms across the Three Subfamilies.Viruses. 2021 Sep 23;13(10):1913. doi: 10.3390/v13101913. Viruses. 2021. PMID: 34696343 Free PMC article. Review.
Cited by
-
A hydrophobic domain within the small capsid protein of Kaposi's sarcoma-associated herpesvirus is required for assembly.J Gen Virol. 2014 Aug;95(Pt 8):1755-1769. doi: 10.1099/vir.0.064303-0. Epub 2014 May 13. J Gen Virol. 2014. PMID: 24824860 Free PMC article.
-
Identification of a Novel, EBV-Based Antibody Risk Stratification Signature for Early Detection of Nasopharyngeal Carcinoma in Taiwan.Clin Cancer Res. 2018 Mar 15;24(6):1305-1314. doi: 10.1158/1078-0432.CCR-17-1929. Epub 2018 Jan 4. Clin Cancer Res. 2018. PMID: 29301829 Free PMC article.
-
A survey of RNA secondary structural propensity encoded within human herpesvirus genomes: global comparisons and local motifs.PeerJ. 2020 Sep 10;8:e9882. doi: 10.7717/peerj.9882. eCollection 2020. PeerJ. 2020. PMID: 32974099 Free PMC article.
-
Epstein-Barr Virus Genomes Reveal Population Structure and Type 1 Association with Endemic Burkitt Lymphoma.J Virol. 2020 Aug 17;94(17):e02007-19. doi: 10.1128/JVI.02007-19. Print 2020 Aug 17. J Virol. 2020. PMID: 32581102 Free PMC article.
-
The assembly domain of the small capsid protein of Kaposi's sarcoma-associated herpesvirus.J Virol. 2012 Nov;86(21):11926-30. doi: 10.1128/JVI.01430-12. Epub 2012 Aug 22. J Virol. 2012. PMID: 22915821 Free PMC article.
References
-
- Addison, C., F. J. Rixon, J. W. Palfreyman, M. O'Hara, and V. G. Preston. 1984. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology 138246-259. - PubMed
-
- Casaday, R. J., J. R. Bailey, S. R. Kalb, E. J. Brignole, A. N. Loveland, R. J. Cotter, and W. Gibson. 2004. Assembly protein precursor (pUL80.5 homolog) of simian cytomegalovirus is phosphorylated at a glycogen synthase kinase 3 site and its downstream “priming” site: phosphorylation affects interactions of protein with itself and with major capsid protein. J. Virol. 7813501-13511. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

