Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India
- PMID: 19158257
- PMCID: PMC2668327
- DOI: 10.1128/JCM.02026-08
Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India
Abstract
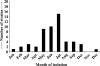
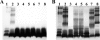
The incidence of Vibrio cholerae non-O1, non-O139 strains from hospitalized patients with acute diarrhea constituted 27.4% (n = 54) of the total 197 V. cholerae strains isolated from patients in Kolkata, India, in 2003. Of 197 strains, 135 were identified as O1 serotype Ogawa and 2 were identified as O139. In the same time period, six O1 background rough strains that possessed all known virulence factors were identified. Serotype analysis of the non-O1, non-O139 strains placed 42 strains into 19 serogroups, while 12 remained O nontypeable (ONT); the existing serotyping scheme involved antisera to 206 serogroups. Detection of a good number of ONT strains suggested that additional serogroups have arisen that need to be added to the current serotyping scheme. The non-O1, non-O139 strains were nontoxigenic except for an O36 strain (SC124), which regulated expression of cholera toxin as O1 classical strains did. Additionally, strain SC124 carried alleles of tcpA and toxT that were different from those of the O1 counterpart, and these were also found in five clonally related strains belonging to different serogroups. Strains carrying tcpA exhibited higher colonization in an animal model compared to those lacking tcpA. PCR-based analyses revealed remarkable variations in the distribution of other virulence factors, including hlyA, rtxA, Vibrio seventh pandemic island I (VSP-I), VSP-II, and type III secretion system (TTSS). Most strains contained hlyA (87%) and rtxA (81.5%) and secreted cytotoxic factors when grown in vitro. Approximately one-third of the strains (31.5%) contained the TTSS gene cluster, and most of these strains were more motile and hemolytic against rabbit erythrocytes. Partial nucleotide sequence analysis of the TTSS-containing strains revealed silent nucleotide mutations within vcsN2 (type III secretion cytoplasmic ATPase), indicating functional conservation of the TTSS apparatus.
Figures


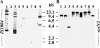
References
-
- Aldova, E., K. Laznickova, E. Stepankova, and J. Lietava. 1968. Isolation of nonagglutinable vibrios from an enteritis outbreak in Czechoslovakia. J. Infect. Dis. 11825-31. - PubMed
-
- Basu, I., R. Mitra, P. K. Saha, A. N. Ghosh, J. Bhattacharya, M. K. Chakrabarti, Y. Takeda, and G. B. Nair. 1999. Morphological and cytoskeletal changes caused by non-membrane damaging cytotoxin of Vibrio cholerae on int 407 and HeLa cells. FEMS Microbiol. Lett. 179255-263. - PubMed
-
- Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45493-496. - PubMed
-
- Bhattacharya, S. K., M. K. Bhattacharya, G. B. Nair, D. Dutta, A. Deb, T. Ramamurthy, S. Garg, P. K. Saha, P. Dutta, A. Moitra, B. K. Mandal, T. Shimada, Y. Takeda, and B. C. Deb. 1993. Clinical profile of acute diarrhoea cases infected with the new epidemic strain of Vibrio cholerae O139: designation of the disease as cholera. J. Infect. 2711-15. - PubMed
-
- Bhattacharya, T., S. Chatterjee, D. Maiti, R. K. Bhadra, Y. Takeda, G. B. Nair, and R. K. Nandy. 2006. Molecular analysis of the rstR and orfU genes of the CTX prophages integrated in the small chromosomes of environmental Vibrio cholerae non-O1, non-O139 strains. Environ. Microbiol. 8526-534. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Medical

