Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation
- PMID: 19158277
- PMCID: PMC2655622
- DOI: 10.1128/MCB.01386-08
Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation
Abstract
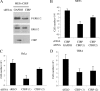
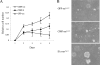
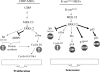
Embryonic stem cells are immortalized cells whose proliferation rate is comparable to that of carcinogenic cells. To study the expression of embryonic stem cell genes in primary cells, genetic screening was performed by infecting mouse embryonic fibroblasts (MEFs) with a cDNA library from embryonic stem cells. Cold-inducible RNA-binding protein (CIRP) was identified due to its ability to bypass replicative senescence in primary cells. CIRP enhanced extracellular signal-regulated kinase 1 and 2 (ERK1/2) phosphorylation, and treatment with an MEK inhibitor decreased the proliferation caused by CIRP. In contrast to CIRP upregulation, CIRP downregulation decreased cell proliferation and resulted in inhibition of phosphorylated ERK1/2 inhibition. This is the first evidence that ERK1/2 activation, through the same mechanism as that described for a Val12 mutant K-ras to induce premature senescence, is able to bypass senescence in the absence of p16(INK4a), p21(WAF1), and p19(ARF) upregulation. Moreover, these results show that CIRP functions by stimulating general protein synthesis with the involvement of the S6 and 4E-BP1 proteins. The overall effect is an increase in kinase activity of the cyclin D1-CDK4 complex, which is in accordance with the proliferative capacity of CIRP MEFs. Interestingly, CIRP mRNA and protein were upregulated in a subgroup of cancer patients, a finding that may be of relevance for cancer research.
Figures






References
-
- Armengol, G., F. Rojo, J. Castellvi, C. Iglesias, M. Cuatrecasas, B. Pons, J. Baselga, and S. Ramon y Cajal. 2007. 4E-binding protein 1: a key molecular “funnel factor” in human cancer with clinical implications. Cancer Res. 677551-7555. - PubMed
-
- Bernard, S. A., T. W. Gray, M. D. Buist, B. M. Jones, W. Silvester, G. Gutteridge, and K. Smith. 2002. Treatment of comatose survivors of out-of-hospital cardiac arrest with induced hypothermia. N. Engl. J. Med. 346557-563. - PubMed
-
- Burdon, T., A. Smith, and P. Savatier. 2002. Signalling, cell cycle and pluripotency in embryonic stem cells. Trends Cell. Biol. 12432-438. - PubMed
-
- Carnero, A., J. D. Hudson, C. M. Price, and D. H. Beach. 2000. p16INK4A and p19ARF act in overlapping pathways in cellular immortalization. Nat. Cell Biol. 2148-155. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
