Characterization of PKD protein-positive exosome-like vesicles
- PMID: 19158352
- PMCID: PMC2637052
- DOI: 10.1681/ASN.2008060564
Characterization of PKD protein-positive exosome-like vesicles
Abstract
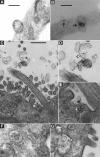
Proteins associated with autosomal dominant and autosomal recessive polycystic kidney disease (polycystin-1, polycystin-2, and fibrocystin) localize to various subcellular compartments, but their functional site is thought to be on primary cilia. PC1+ vesicles surround cilia in Pkhd1(del2/del2) mice, which led us to analyze these structures in detail. We subfractionated urinary exosome-like vesicles (ELVs) and isolated a subpopulation abundant in polycystin-1, fibrocystin (in their cleaved forms), and polycystin-2. This removed Tamm-Horsfall protein, the major contaminant, and subfractionated ELVs into at least three different populations, demarcated by the presence of aquaporin-2, polycystin-1, and podocin. Proteomic analysis of PKD ELVs identified 552 proteins (232 not yet in urinary proteomic databases), many of which have been implicated in signaling, including the molecule Smoothened. We also detected two other protein products of genes involved in cystic disease: Cystin, the product of the mouse cpk locus, and ADP-ribosylation factor-like 6, the product of the human Bardet-Biedl syndrome gene (BBS3). Our proteomic analysis confirmed that cleavage of polycystin-1 and fibrocystin occurs in vivo, in manners consistent with cleavage at the GPS site in polycystin-1 and the proprotein convertase site in fibrocystin. In vitro, these PKD ELVs preferentially interacted with primary cilia of kidney and biliary epithelial cells in a rapid and highly specific manner. These data suggest that PKD proteins are shed in membrane particles in the urine, and these particles interact with primary cilia.
Figures







References
-
- Torres V, Holley K, Offord K. Epidemiology. In: Problems in Diagnosis and Management of Polycystic Kidney Disease, edited by Grantham JJ, Kansas City: PKR Foundation Kansas City, 1985, pp 349–369
-
- Wilson PD: Polycystic kidney disease. N Engl J Med 350: 151–164, 2004 - PubMed
-
- The polycystic kidney disease 1 gene encodes a 14 kb transcript and lies within a duplicated region on chromosome 16. The European Polycystic Kidney Disease Consortium. Cell 77: 881–894, 1994 - PubMed
-
- Polycystic kidney disease: The complete structure of the PKD1 gene and its protein. The International Polycystic Kidney Disease Consortium. Cell 81: 289–298, 1995 - PubMed
-
- Hughes J, Ward CJ, Peral B, Aspinwall R, Clark K, Millan JL, Gamble V, Harris PC: The polycystic kidney disease 1 (PKD1) gene encodes a novel protein with multiple cell recognition domains. Nat Genet 10: 151–160, 1995 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

