The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly
- PMID: 19158390
- PMCID: PMC2655267
- DOI: 10.1091/mbc.e08-08-0838
The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly
Abstract
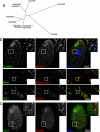
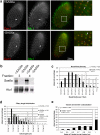
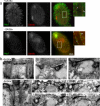
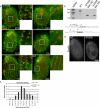
Cilia and flagella are structurally and functionally conserved organelles present in basal as well as higher eukaryotes. The assembly of cilia requires a microtubule based scaffold called a basal body. The ninefold symmetry characteristic of basal bodies and the structurally similar centriole is organized around a hub and spoke structure termed the cartwheel. To date, SAS-6 is one of the two clearly conserved components of the cartwheel. In some organisms, overexpression of SAS-6 causes the formation of supernumerary centrioles. We questioned whether the centriole assembly initiation capacity of SAS-6 is separate from or directly related to its structural role at the cartwheel. To address this question we used Tetrahymena thermophila, which expresses two SAS-6 homologues, TtSAS6a and TtSAS6b. Cells lacking either TtSAS6a or TtSAS6b are defective in new basal body assembly. TtSas6a localizes to all basal bodies equally, whereas TtSas6b is enriched at unciliated and assembling basal bodies. Interestingly, overexpression of TtSAS6b but not TtSAS6a, led to the assembly of clusters of new basal bodies in abnormal locations. Our data suggest a model where TtSAS6a and TtSAS6b have diverged such that TtSAS6a acts as a structural component of basal bodies, whereas TtSAS6b influences the location of new basal body assembly.
Figures



References
-
- Badano J. L., Mitsuma N., Beales P. L., Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–148. - PubMed
-
- Brown T. Southern blotting. In: Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. Current Protocols in Molecular Biology. Vol. 1. Hoboken, NJ: John Wiley and Sons; 1999. pp. 2.9.1–2.9.15.
-
- Bruns P. J., Cassidy-Hanley D. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 2000;62:501–512. - PubMed
-
- Callaini G., Whitfield W. G., Riparbelli M. G. Centriole and centrosome dynamics during the embryonic cell cycles that follow the formation of the cellular blastoderm in Drosophila. Exp. Cell Res. 1997;234:183–190. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

